Produced in Partnership with XenocsDec 2 2020
3D envelopes, including three different proteins with low volume and dilute solutions, were established via an automated sequence of measurements.
Introduction
Small-angle X-ray scattering (SAXS) is becoming an important part of structural biology techniques. It provides the appropriate information regarding the shape of proteins in solution and any associated parameters, including dynamics and macromolecule interactions. The reliability and high throughput of the measurements are crucial requirements for the capacity to effectively run samples in a structural biology laboratory.
This article outlines the quality of three-dimensional envelopes amassed on three protein samples (bovine serum albumin (BSA), lysozyme, and thyroglobulin) utilizing an automated measurement sequence, measured on the BioXolver.
Measurements & results
Concentration series were performed, using the in-line pipetting robot, which enables automate sample loading and cell cleaning in a cycle time, which is under two minutes. Three concentrations successively ran per sample, and for the measurements, 5 µL per sample condition were employed.
To determine the quality of the sample, running treatment of the data through the automated software package based on ATSAS1 facilitates rapid calculations of the corresponding 3D envelopes for each concentration.
The entire sequence of measurements was concluded in under 90 minutes, including the three concentrations per buffer and sample measurements: including from the loading of the well plate through to the generation of the final 3D envelopes. To highlight the high quality of the data, further data processing was conducted using ATSAS.
The 3D protein envelopes generated by DAMMIN2 for the lowest measured concentrations of BSA are exhibited in Figures 1 and 2. By covering the homologous crystal structure for each of the samples, collation of the calculated shape to existing crystallographic data is provided. Comparative analysis demonstrates excellent consistency of the approximated shape.
This determines the capacity to establish 3D envelopes from weakly scattering samples in short measurement time (five minutes exposure time). Comparative analysis between the synchrotron SAXS data and BioXolver measurements for the thyroglobulin sample are shown in Figure 3.
Duplication of the analysis procedure, which produced DAMMIF/DAMMIN models, was applied to both SAXS data sets. The resultant 3D envelopes are extraordinarily similar and demonstrate the high quality of the results acquired using the BioXolver in a five minutes exposure time.
Conclusion
The BioXolver’s capacity to yield high-quality results with a reliable, high throughput while applying small volumes of protein solution is observable in the results. It sets the scene for routine characterization of biological macromolecules in the laboratory, providing rapid feedback for formulation studies and other research.
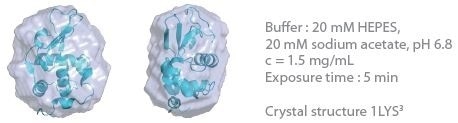
Figure 1. 3D envelope from a lysozyme solution (Mw = 14.3 kDa). Image Credit: Xenocs
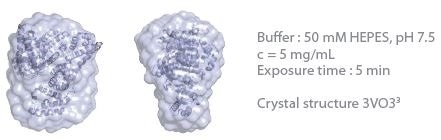
Figure 2. 3D envelope from a BSA solution (Mw = 66 kDa). Image Credit: Xenocs
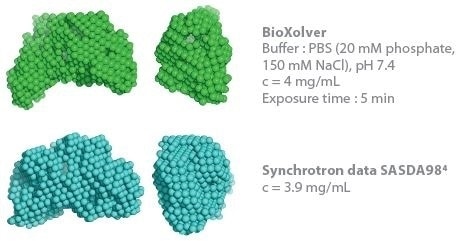
Figure 3. Comparison of 3D envelopes of thyroglobulin (Mw = 669 kDa). Image Credit: Xenocs
References
- D. Franke et al. (2017) J. Appl. Cryst. 50
- D. I. Svergun (1999), Biophys J. 2879-2886.
- http://www.rcsb.org
- http://www.sasbdb.org
About Xenocs
Xenocs is a supplier of x-ray scattering equipment (SAXS/WAXS) for characterizing the nanostructure and morphology of materials at the nanoscale. Such equipment is used for the research, development, and production of advanced materials in various domains such as nanoparticles & colloïds, polymer, food science, cosmetics, or biostructural research.
Created in 2000 as a spin-off from Institute Laue Langevin, the company started activity offering its customers innovative X-ray sources, optics, and collimation solutions for X-ray characterization of nanomaterials and nanostructures.
Xenocs delivered its first x-ray scattering equipment in 2008, setting new standards for the possibilities of using SAXS in the laboratory for characterization at the nanoscale.
Working closely with both academic and corporate customers, Xenocs has continuously focused on providing value through performance and ease of use, while also creating a global sales and service organization.
In 2016, Xenocs bought Saxslab with operations in Denmark and in Massachusetts, making it the leading provider of SAXS equipment worldwide. Xenocs headquarters are located in France, and the company has subsidiaries in the USA, Denmark, and Singapore as well as a strong network of local contacts.
Sponsored Content Policy: AZO Life Science publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of AZO Life Science, which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.