Examining the gene expression of a person needs mapping the RNA landscape to a standard reference. This is done to gain an understanding of the degree to which genes are “turned on” and execute the functions in the body.
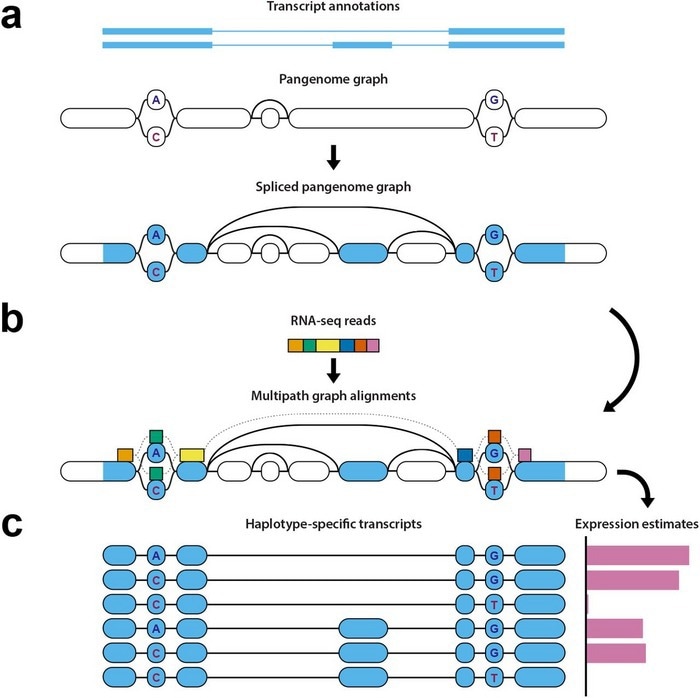 Diagram of haplotype-aware transcriptome analysis pipeline. Image Credit: Credit to study authors.
Diagram of haplotype-aware transcriptome analysis pipeline. Image Credit: Credit to study authors.
However, scientists can face issues when the reference does not offer sufficient information to enable precise mapping, a problem called reference bias.
In a new study performed, scientists at the University of California Santa Cruz (UC Santa Cruz) introduce the first-ever technique for examining RNA sequencing data genome-wide by making use of a “pantranscriptome.”
This integrates a transcriptome and a pangenome—a reference that comprises genetic material from a cohort of different individuals, instead of just a single linear strand.
The study has been reported in the Nature Methods journal
A research group headed by UCSC Associate Professor of Biomolecular Engineering Benedict Paten has released a toolkit. This enables scientists to map an individual’s RNA data to a much wealthier reference. It fulfills reference bias and results in much more precise mapping.
This is pangenome plus transcriptome—that combination has never really been done before until now. This is the first time anyone has attempted to incorporate the pangenome as a standard feature of the RNA sequencing mapping.”
Jordan Eizenga, Study Co-First Author and Postdoctoral Scholar, Computational Genomics Lab, University of California-Santa Cruz
Globally, this newly-developed tool will help scientists who are working to comprehend gene expression via RNA sequencing analysis. The tools are available in public and can also be accessed through Github.
With this toolkit, we are employing this more diverse data that we can now get from the pangenome to improve the measurement of gene expression data, something that can widely vary between individuals.”
Benedict Paten, Associate Professor, Biomolecular Engineering, University of California-Santa Cruz
Paten added, “The aim is to make the impact of this more diverse data felt on studies that are looking at gene expression, resulting in better analysis for cell models, organoid models, and other research applications.”
The most generally identified function of RNA is to translate DNA into proteins. However, researchers currently have understood the fact the huge majority of RNA seems to be noncoding and does not make proteins, but rather could play roles like impacting cell structure or controlling genes.
The complete RNA landscape is known jointly as the transcriptome, and mapping this enables scientists to better comprehend a gene expression of the individual.
The pantranscriptome relies on the rising concept of “pangenomics” in the genomics field. Normally, while assessing the genomic data of an individual for variation, researchers make a comparison of the individual’s genome to that of a reference fabricated of a single linear strand of DNA bases.
Making use of a pangenome enables scientists to make a comparison of an individual’s genome to that of a genetically different cohort of reference sequences all right away. This has been sourced from individuals symbolizing a diversity of biogeographic ancestry. This offers the researchers more points of comparison for which to better comprehend a genomic variation of the individual.
It can be hard to comprehend gene expression by mapping the RNA sequencing data as the RNA sequences are spliced by cellular mechanisms. This implies that one set of RNA data can come from the genome’s non-connected areas, thereby making it hard to align them properly to a reference.
Such splicing sites are not even throughout the human population but change between individuals. Also, it is hard to identify which haplotype the RNA comes from—if the group of genes comes particularly from the set of chromosomes that have been inherited from the individual’s mother, or the set inherited from the father.
However, with the new pipeline of open-source tools, scientists can take the spliced segments of an individual’s RNA, then map where they align on a pangenome, determine which haplotype the data belongs to, and examine gene expression.
Initially, the pipeline determines which areas of the genome the RNA sequencing data comes from, such as the splice sites, and signs those points on the pangenome reference.
Furthermore, those marked points are compared to a pantranscriptome comprising haplotype-specific transcripts produced from the reference data contained inside the pangenome. This step needs specialized and difficult algorithmic techniques.
Eventually, it produces estimates of levels of gene expression depending on this comparison between the mapped data and then the transcripts in the pantranscriptome, and determines which haplotypes the genes come from.
It's definitely a very forward-looking study in that other genome-wide expression methods are not yet really utilizing pangenomes and haplotype information. We're now thinking ahead as to what pangenomics might additionally bring to the table in transcriptomic analyses.”
Jonas Sibbesen, Study Co-First Author and Former Postdoctoral Scholar, Computational Genomics Lab, University of California-Santa Cruz
At present, Sibbesen is working as an assistant professor at the University of Copenhagen.
Going ahead, the scientists are interested in additional developing such tools to be beneficial for downstream informatics analysis. Also, it customizes the tools for the particularities of study on single-cell data. Currently, the group believes their new toolkit will serve to exhibit how beneficial using pangenomics-derived analysis could be.
Paten stated, “We need to be able to explain to some researchers how a pangenome reference will benefit them. This pipeline is really a first go at doing this for RNA, for functional data, for expression data.”
Source:
Journal reference:
Sibbesen, J. A., et al. (2022) Haplotype-aware pantranscriptome analyses using spliced pangenome graphs. Nature Methods. doi.org/10.1038/s41592-022-01731-9.