The quantity of structural data available on protein drug targets keeps growing. Nevertheless, efficiently mining this information to create testable hypotheses that influence drug discovery can be tough.
Hotspot maps use empirical data to assess protein binding sites to understand the druggability of the pocket, prioritize drug design, and spot differences in similar proteins that might drive compound selectivity. Image Credit: Cambridge Crystallographic Data Centre
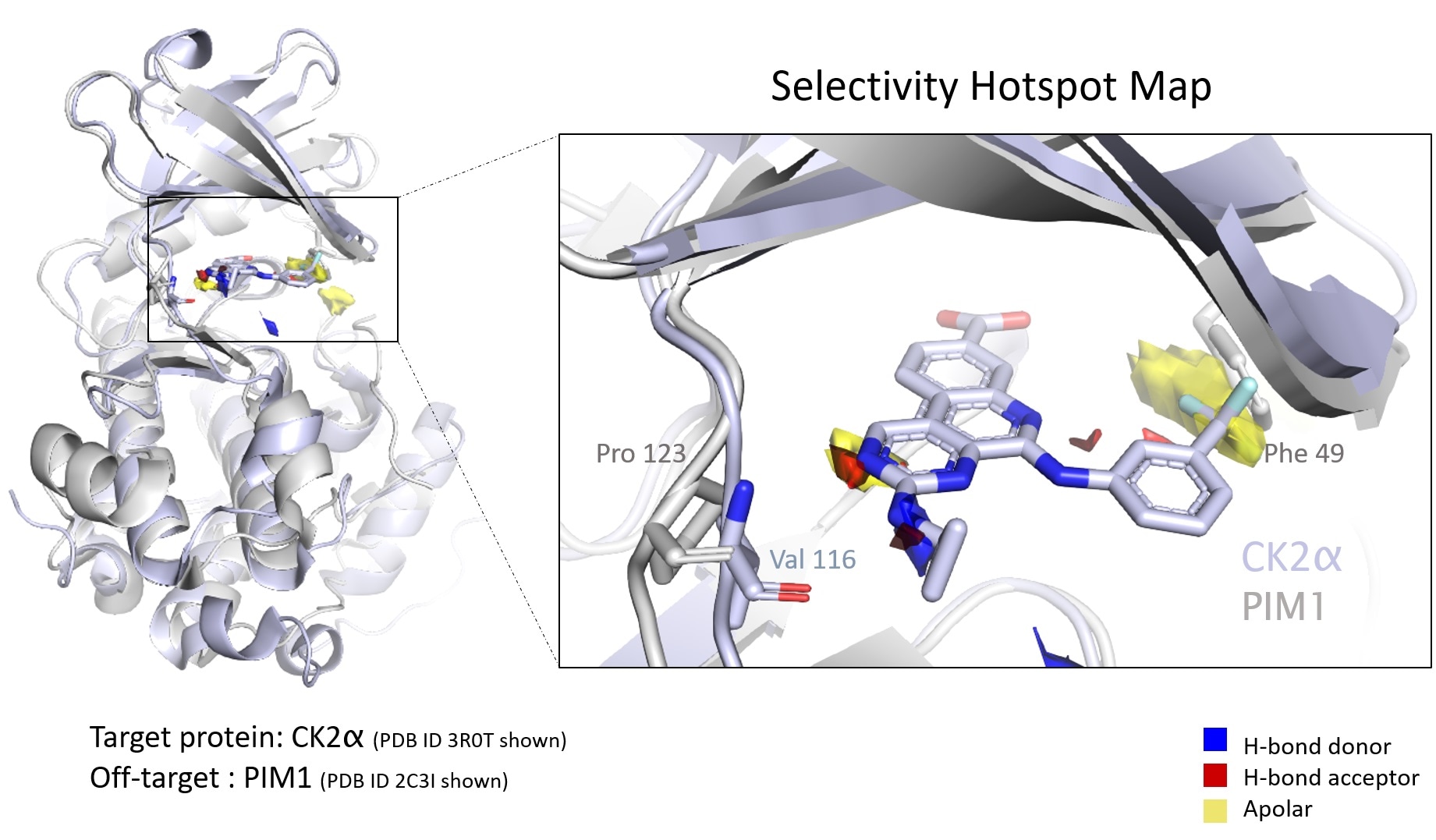
The target protein’s selectivity is a critical property in the designing of new treatments. In a recent study, authors from the CCDC, Exscientia, and Oxford University reveal how an automatic approach leveraging “ensemble hotspot maps” can detect vital structural differences that attribute to the compound’s selectivity for one protein over another. The paper was published in the Journal of Chemical Information and Modeling.
The power of hotspot mapping to advance drug design
Hotspot mapping measures the propensity for compounds in a preferred binding site to exploit interactions—offering a 3D grid of data to aid in scoring and prioritizing compounds. The strength of this method depends on how it discovers vital interactions in early-phase drug discovery and then filters the data into results that are easily interpretable.
Adding hotspot maps early in a drug discovery project can provide a molecular blueprint using the protein structure alone. This can be used to help determine how druggable a given pocket of a target protein is and to prioritize fragment starting points for compound design. The highest scoring interactions can then be used to guide computational methods and algorithms.”
Chris Radoux, Study Co-Author and Head, Structural Bioinformatics, Exscientia
Hotspot maps drive cohesive drug design
This method automates analysis throughout a protein family since proteins present in the same family usually have identical binding sites.
As per Mihaela D Smilova, co-author and postgraduate researcher at the Centre for Medicines Discovery at Oxford University, selectivity profiles present in both the complete proteome—and in the target protein family—need to be comprehended to make effective and safe drugs.
She adds that contacts with dissimilar target proteins can result in toxicity and unnecessary side effects. However, successful drugs abuse the advantages of “polypharmacology” often.
Introducing polypharmacology, or the ability to modulate multiple targets, may help to prevent the development of resistant disease phenotypes. Consequently, a successful drug candidate has a finely tuned selectivity profile within its target family—interacting with targets that positively impact the disease phenotype and avoiding interactions that lead to unwanted side effects.”
Mihaela D Smilova, Study Co-Author and Postgraduate Researcher, Centre for Medicines Discovery, Oxford University
Using inputs in the form of hotspot maps for computational workflows implies scientists can quickly discover the chemical space.
“This saves time by summarizing the information and presenting it in a way that is both interpretable by medicinal chemists and can be used in further computational analyses,” states Smilova.
Leveraging real-world, empirical data for reliability
A Python package—called Hotspots API—is the script used to produce the hotspot maps. This leverages the information in the Cambridge Structural Database (CSD) through CCDC’s IsoStar library of interactions. The CSD is the repository of the world for small-molecule organic and metal-organic crystal structures. It contains more than 1.1 million structures from neutron diffraction analyses and x-ray.
IsoStar—a web application—uses the CSD to produce thousands of interactive 3D scatterplots that reveal the occurrence probability and spatial features of interactions among pairs of chemical functional groups.
Using CSD data for this type of analysis provides different insights from energy-calculation-based methods, as the interactions observed in the CSD are influenced by more than their strength.”
Dr Jason Cole, Senior Research Fellow, Cambridge Crystallographic Data Centre
Impacts of the study
Exscientia, which is present at the interface of sophisticated AI applications and complicated drug discovery, is an international leader in pharmatech. It has employed the hotspot mapping in-house in several drug discovery programs and utilized it to direct target evaluation and drug development.
Also, a research group at the University of Cambridge has recently published how they employed fragment hotspot mapping to detect structures that may help in developing DNA-dependent protein kinase catalytic subunit inhibitors that are showing possibilities as cancer medicines.
Source:
Journal reference:
Smilova, M. D., et al. (2022) Fragment Hotspot Mapping to Identify Selectivity-Determining Regions between Related Proteins. Journal of Chemical Information and Modeling. https://doi.org/10.1021/acs.jcim.1c00823.