This article is based on a poster originally authored by SungHee Park, Andrew Bond, George Maio, and Li-Tao Guo.
Reverse transcription combined with template switching offers a reliable and efficient method for generating full-length cDNA libraries and enabling adapter ligation in RNA-Seq workflows.
However, traditional protocols that rely on Moloney murine leukemia virus (MMLV) reverse transcriptases (RTs) often fall short, especially when dealing with the complex RNA structures and lengths commonly found in plant and agricultural samples.
Many RNA transcripts central to plant biology, such as those linked to stress responses, pathogen resistance, and regulatory networks, are both long and structurally intricate. These characteristics make them difficult to process using standard RT enzymes, which frequently stall during transcription. As a result, important transcripts may go undetected, limiting the effectiveness of transcriptomic analyses in agricultural research.
In response to these obstacles, this article presents the UltraMarathonRT® (uMRT) enzyme, an ultra-processive RT that can maintain high fidelity over long and structured RNA sequences.
In this article, the researchers describe the enzyme's involvement in improved library preparation for transcriptomics and compare it to standard MMLV-based approaches.
The researchers reduced non-specific amplification of RT primers and template-switching oligonucleotides (TSO), which can conceal transcript representation in various plant RNA samples, using systematic optimization.
Using uMRT, RNA-Seq was performed on a well-characterized RNA reference set and a variety of RNA samples, resulting in sequencing reads that reliably captured RNA identity and abundance throughout a six-log range of transcript concentrations.
This increased sensitivity and accuracy are crucial for plant research applications because they enable the discovery of uncommon transcripts and better representation of full-length mRNAs.
The researchers also confirmed the efficacy of this method using whole RNA samples, exposing a broader spectrum of transcripts, including those previously undetectable by standard RT enzymes.
In practice, the uMRT enzyme facilitates plant research by allowing for thorough transcriptome assessments of stress responses, pathogen interactions, and developmental regulations, which are critical for increasing crop resilience and productivity.
This RNA-Seq pipeline, which uses advanced enzymology and optimizes RT performance, represents a significant step forward in plant and agricultural transcriptomics, allowing for comprehensive profiling of complex RNA populations and supporting discoveries that could inform crop improvement strategies and sustainable agricultural practices.
UltraMarathonRT: Unmatched Processivity for Long, Structured RNA
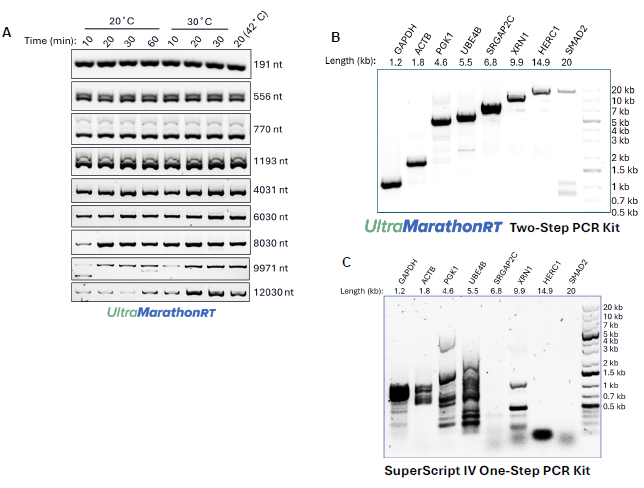
Figure 1. Sensitive, unbiased full-length cDNA synthesis from RNA of any length and structure content by uMRT. (A) RT-PCR amplification of target RNA templates of varying lengths at different temperatures. The target RNA templates were amplified from 10 pg SIRVSet 4 (Lexogen, SKU 141.01) spike-in RNAs. (B) and (C) Comparing the amplification of human RNAs with lengths ranging from 1.2 kb to 20 kb using (B) uMRT Two-Step RT-PCR Kit (RNAConnect, Cat#R1005) and (C) SuperScript IV One-Step RT-PCR Kit (Thermo, Cat#12594025). Reverse transcription for both reactions was performed using 100 ng of Hela total cellular RNA (Takara, Cat#636543). All other aspects of RT and PCR reaction procedures for SuperScript IV were followed according to their respective manufacturer recommendations (see reference 1). Image Credit: RNA Connect
UltraMarathonRT Enables More Comprehensive Profiling of Human Transcriptome by Improving the Visibility of Difficult RNAs
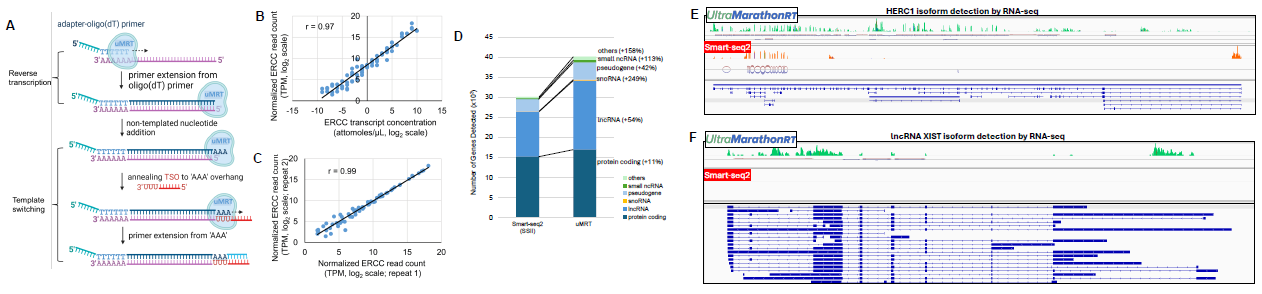
Figure 2. RNA-seq benchmarking using universal human reference RNA (Thermo, Cat# QS0639). (A) Outline of template switching mediated cDNA synthesis from poly(rA) RNA by uMRT. (B) Linear regression showing excellent correlation between ERCC RNA transcript concentration (x-axis) used in library preparation and the normalized read count (TPM) (y-axis) calculated from the sequencing reads. The TPM values and ERCC transcript concentrations were log2 transformed. (C) Examining the reproducibility of RNA-seq experiment by comparing the normalized ERCC read count (log2 transformed TPM) of two repeated experiments (see reference 2). (D) Bar charts illustrating the number of genes detected (TPM > 0) between uMRT and Smart-seq (using SSII), respectively, with reads downsampled to 6 million each sample to
account for sequencing depth. Colors indicate the gene biotype of the detected genes, depicting the distribution across all detected biotypes. (E) IGV browser tracks showing HERC1 detection in the RNA-seq experiment. The comparison shows that uMRT, rather than Smart-seq2, detects the full-length long isoform, which agrees well with the RT-PCR result in Figure 1B. (F) IGV browser tracks showing XIST lncRNA detection by uMRT, rather than Smart-seq2, in the RNA-seq experiment. Image Credit: RNA Connect
UltraMarathonRT Accurately Captures the Transcriptomic Profiles and Gene Expression Levels of Rice RNA
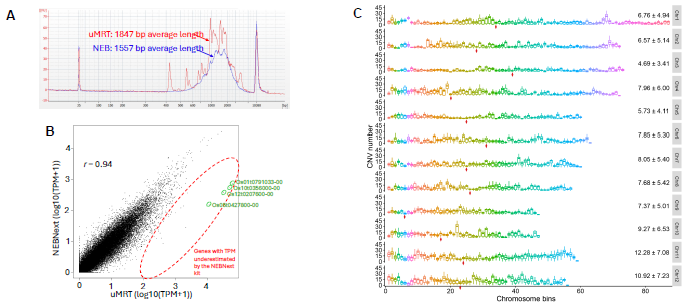
Figure 3. RNA-seq benchmarking using rice leaf total RNA (Zyagen, Cat# PLR-1004). The rice reference genome and gtf annotations used in the analysis were downloaded from The Rice Annotation Project Database (RAP-DB). (A) BioAnalyzer profiles of pre-amplified full-length cDNA libraries generated by uMRT template switching kit (RNAConnect, Cat# R1004S) and NEBNext® Single Cell/Low Input cDNA Synthesis & Amplification Module (NEB, Cat# E6421S). The two profiles were superimposed to highlighted their difference. (B) Scatterplot comparison of TPM normalized gene counts (log10(TPM+1) scale) between samples prepared with uMRT and the NEB kits. The genes that show highly differentiated TPM between uMRT and the NEBNext datasets were highlighted in red circle (see Table 2 for more details about the four named transcripts highlighted in green circles). (C) The CNV (Copy number variation) numbers of 500-kb non-overlap windows at 12 rice chromosomes (adapted from reference 3). The red diamonds marked the positions of the centromere. Each boxplot represents the CNV number of the corresponding location in 93 rice genome accessions. Image Credit: RNA Connect
Table 1. Summary of RNA-seq reads mapped to duplicated genes. Rice genome is highly duplicated (Figure 3C), and the uMRT dataset excellently captures this feature. Source: RNA Connect
Count of
duplication |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
>10 |
| uMRT |
39.5 % |
3.4 % |
4.8 % |
5.3 % |
6.6 % |
8.7 % |
8.2 % |
7.0 % |
8.5 % |
2.2 % |
5.8 % |
| NEBNext |
90.5 % |
1.2 % |
0.6 % |
0.4 % |
0.6 % |
0.8 % |
0.9 % |
0.9 % |
0.7 % |
0.3 % |
3.2 % |
Table 2a. Representative genes/transcripts that show drastically different expression levels (TPM) between uMRT and NEBNext datasets. The four transcripts highlighted in green were labeled in Figure 3B. These four transcripts encode the same protein (RuBisCO large subunit), which is the critical enzyme for photosynthesis and photorespiration and is expected to be highly expressed in leaves. RT-qPCR result confirmed this (see Table 2b). Source: RNA Connect
| Transcript ID |
Chr |
TPM |
Gene |
| uMRT |
NEB |
| Os01t0791033-00 |
1 |
62155.5 |
732.3 |
RBCL1(photosynthesis and photorespiration) |
| Os05t0427800-00 |
5 |
11421.4 |
148.2 |
| Os10t0356000-00 |
10 |
50942.1 |
548.9 |
| Os12t0207600-00 |
12 |
32811.0 |
368.1 |
| Os01t0885200-01 |
1 |
124.4 |
5.6 |
Putative G protein-coupled receptor |
| Os02t0148000-01 |
2 |
114.9 |
12.4 |
CCT domain containing protein |
| Os03t0268050-00 |
3 |
111.2 |
9.3 |
Non-protein coding transcript |
| Os03t078100-01 |
3 |
58.7 |
31.1 |
ACT1 |
| Os04t0390000-01 |
4 |
125.6 |
9.9 |
alpha-ketoglutarate dehydrogenase (key enzyme in TAC cycle) |
| Os05t0530500-03 |
5 |
152.0 |
10.6 |
Serine/threonine protein kinase |
| Os06t0558700-01 |
7 |
134.4 |
14.5 |
Poly(A) polymerase, RNA-binding region domain containing protein |
| Os08t0433350-00 |
8 |
217.2 |
13.1 |
photosystem II protein I |
| Os09t0553700-01 |
9 |
106.5 |
14.8 |
Similar to PRLI-interacting factor A |
| Os10t0355800-00 |
10 |
600.2 |
73.3 |
ATP synthase CF1 beta subunit |
| Os11t0246900-02 |
11 |
142.5 |
13.6 |
Ankyrin repeat-containing domain superfamily |
| Os12t0568750-00 |
12 |
185.2 |
13.5 |
Hypothetical protein |
Table 2b. RT-qPCR validation of gene expression for RNA-seq results. Source: RNA Connect
| RT-qPCR validation |
| Gene: Os10t0356000-00 (RBCL1) |
| uMRT RT-qPCR (Ct) |
NEB Luna One-Step (Ct) |
| 10.7 ± 0.02 |
9.9 ± 0.06 |
| Gene: Os03t078100-01 (ACT1) |
| uMRT RT-qPCR (Ct) |
NEB Luna One-Step (Ct) |
| 22.1 ± 0.3 |
21.7 ± 0.09 |
Note: UltraMarathonRT Two-Step RT-qPCR Kit (RNAConnect, ref#R1006) and Luna Universal One-Step RT-qPCR Kit (NEB, ref#E3005) were used to validate the RNAseq results. Both kits show the RBCL1 gene is highly expressed, confirming the accuracy of uMRT RNA-seq results. ACT1 gene was used as the internal control.
Conclusion
- UltraMarathonRT offers complete coverage of any RNA transcript, regardless of sequence, structure, or length.
- UltraMarathonRT outperformed SuperScript II, an MMLV RT, in capturing long, structured RNAs produced in the human transcriptome.
- UltraMarathonRT accurately recreated rice gene duplication and expression levels.
References
- Thermo Fisher Scientific Inc. (2017). USER GUIDE Pub. No. MAN0016518 rev. A.0 SuperScriptTM IV One-Step RT-PCR System. Available at: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0016518_superscriptIV_1step_master_mix_UG.pdf.
- Guo, L.-T., et al. (2024). Characterization and implementation of the MarathonRT template-switching reaction to expand the capabilities of RNA-Seq. RNA, (online) 30(11), pp.1495–1512. https://doi.org/10.1261/rna.080032.124.
- Zhao, F., et al. (2020). A genome-wide survey of copy number variations reveals an asymmetric evolution of duplicated genes in rice. BMC Biology, 18(1). https://doi.org/10.1186/s12915-020-00798-0.
About RNA Connect
RNAConnect was founded by innovators in the RNA community for innovators in the RNA community. We think of enzymes as “biotechnology hardware,” and we believe that better enzymes provide more accurate and complete visibility into RNA which allows you to discover, develop, and innovate faster, easier and in ways not previously possible.
Sponsored Content Policy: AZO Life Sciences publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of AZO Life Sciences which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Oct 10, 2025