Reviewed by Danielle Ellis, B.Sc.Aug 30 2023
Researchers discovered a new type of marine bacteria with distinct traits on the ocean floor.
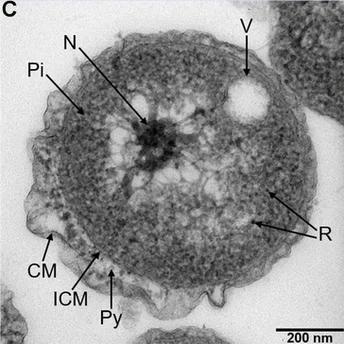 The novel bacteria, which the team proposes to name Poriferisphaera hetertotrophicis, observed using Transmission Electron Microscopy (TEM). Abbreviations: CM, outer membrane; Pi, cytoplasm; R, ribosome; N, nucleoid; ICM, cytoplasmic membrane; Py, periplasm; V, vesicle-like organelles. Image Credit: Rikuan Zheng
The novel bacteria, which the team proposes to name Poriferisphaera hetertotrophicis, observed using Transmission Electron Microscopy (TEM). Abbreviations: CM, outer membrane; Pi, cytoplasm; R, ribosome; N, nucleoid; ICM, cytoplasmic membrane; Py, periplasm; V, vesicle-like organelles. Image Credit: Rikuan Zheng
The study, which is described by the editors as a significant one that advances the current understanding of physiological mechanisms in deep-sea Planctomycetes bacteria, reveals unusual characteristics like being the only known species in the class of Phycisphaerae bacteria that employs a distinct budding model of division and was published on August 29th, 2023 as a Reviewed Preprint in eLife.
It also offers what the editors describe as compelling proof that the novel species is heavily invested in nitrogen absorption and coexists with a persistent virus (bacteriophage) that promotes nitrogen metabolism. Nitrogen cycling by bacteria is a crucial mechanism that releases nitrogen for use in the synthesis of nucleic acids, amino acids, and proteins, which are the building blocks of life.
Until recently, most research on the Planctomycetes family of bacteria has focused on strains in freshwater and shallow ocean environments, because of the logistical difficulties associated with sampling and cultivating deep-sea strains. Most Planctomycetes bacteria have been isolated using growth media that are nutritionally poor, so we wanted to see if using a nutrient-rich medium would make it possible to culture and further characterize members of this poorly understood family.”
Rikuan Zheng, Study Lead Author and Research Associate, Institute of Oceanology, Chinese Academy of Sciences
The scientists used sediment samples from a deep-sea cold seep, where Planctomycetes bacteria are known to exist, to identify the unknown bacterium. They then promoted their development by adding rifampicin and nitrogen sources to a conventional growing medium. They cultivated these enhanced bacteria on agar and performed gene sequencing on certain colonies to assess them further.
Image Credit: Kateryna Kon/Shutterstock.com
They discovered a strain of bacteria they dubbed ZRK32 that grew more quickly than the others and appeared to belong to the genus Poriferisphaera. This was supported by a comparison of the genetic similarity between this strain and other Poriferisphaera genus members, which revealed that it could be distinguished from Poriferisphaera corsica, the only other species having a recognized published name. The team advises naming ZRK32 Poriferisphaera hetertotrophicis in light of the evidence that ZRK32 represents a distinct species.
To learn more about this new species, the team studied its growth and how it multiplies. They found that, unlike other Planctomycetes family members, Poriferisphaera hetertotrophicis grows better in nutrient-rich media and multiplies via a budding mechanism, where parent cells create outgrowth buds that develop into daughter cells.
The scientists then investigated if Poriferisphaera hetertotrophicis shared the Planctomycetes bacteria family’s crucial function in nitrogen cycling. They examined how the development of Poriferisphaera hetertotrophicis was affected by nitrates, ammonia, and nitrogen dioxide to test this. They discovered that supplying nitrogen as nitrite reduced growth while adding it as nitrate enhanced growth.
The researchers next investigated if this was also true for Poriferisphaera hetertotrophicis since the Planctomycetes bacteria family is known to play a significant role in nitrogen cycling.
They examined how the development of Poriferisphaera hetertotrophicis was affected by several nitrogen-containing substances, including nitrates, ammonia, and nitrogen dioxide, to test this. They discovered that providing nitrogen in the form of ammonia or nitrate promoted growth while adding nitrite prevented it.
They also found that the unique strain released a bacteriophage, a type of virus that infects bacteria, in response to the addition of nitrate or ammonia. Oceans are home to a large population of bacteriophages, which can control the metabolism of their host bacteria’s nitrogen.
By improving nitrogen metabolism, the virus, also known as phage-ZRK32, was able to significantly boost Poriferisphaera hetertotrophicis and other marine bacteria's rate of growth.
Poriferisphaera hetertotrophicis could include all the required genes for metabolizing nitrate and ammonia, according to the team’s genetic investigation, but prolonged infection with this bacteriophage could help to further optimize nitrogen metabolism.
Our analyses indicate that strain ZRK32 is a novel species, which grows best in nutrient-rich media and releases a bacteriophage in the presence of nitrogen. This phage-ZRK32 is a chronic bacteriophage that lives within its host without killing it. Our findings provide a novel insight into nitrogen metabolism in Planctomycetes bacteria and a suitable model to study the interactions between Planctomycetes and viruses.”
Chaomin Sun, Study Senior Author and Professor, Institute of Oceanology, Chinese Academy of Sciences
Source:
Journal reference:
Zheng, R., et al. (2023). Physiological and metabolic insights into the first cultured anaerobic representative of deep-sea Planctomycetes bacteria. eLife. doi.org/10.7554/eLife.89874.1.