Reviewed by Lexie CornerMay 20 2025
Researchers at the Francis Crick Institute and the Gulbenkian Institute for Molecular Medicine (GIMM) have discovered that the evolution of a family of exported proteins in the malaria-causing parasite Plasmodium falciparum enabled it to infect humans.
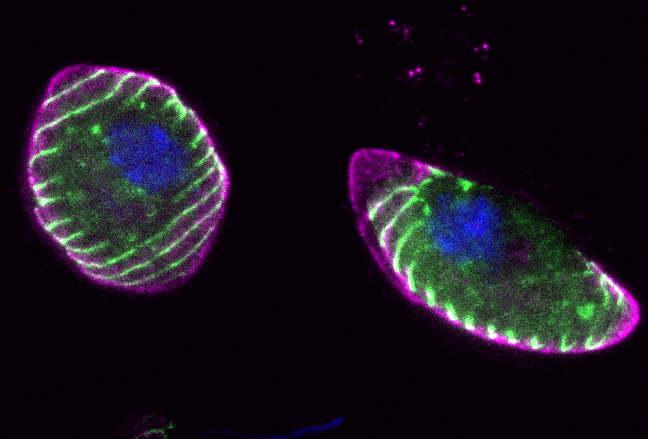 Immunofluorescence image showing FIKK kinase (green) inside gametocytes (magenta, the transmissible form of the parasite). Image Credit: Hugo Belda and Crick Advanced Light Microscopy team
Immunofluorescence image showing FIKK kinase (green) inside gametocytes (magenta, the transmissible form of the parasite). Image Credit: Hugo Belda and Crick Advanced Light Microscopy team
Targeting these proteins may lead to the development of new drugs that are less susceptible to resistance.
Malaria affects approximately 200 million people and causes over 500,000 deaths each year. It is caused by Plasmodium parasites that invade red blood cells and spread between hosts through mosquito bites. While treatments exist, drug resistance remains a persistent challenge.
In a study published today in Nature Microbiology, a team of malaria researchers at the Crick - now based at the Gulbenkian Institute for Molecular Medicine (GIMM) - investigated the most lethal Plasmodium species, P. falciparum, which is responsible for more than 95 % of malaria-related deaths.
Parasitic Tools to Target Human Cells
After a malaria infection, the P. falciparum parasite exports about 10% of its proteins into host red blood cells, causing them to stick to blood vessels and to each other, which can lead to blood clots.
Among these exported proteins is a family known as FIKK kinases, which modify host molecules or other parasite-exported proteins to regulate their activity.
The researchers analyzed more than 2,000 P. falciparum samples from malaria patients and found that 18 of the 21 FIKK kinases were highly conserved and protected from harmful mutations. This suggests they are essential for human infection and likely played a role in the parasite’s adaptation to humans.
To investigate their function, the team expressed the FIKK kinases in bacteria and tested their activity. The study showed that each kinase had specific protein targets within the host cell.
Unexpectedly, the researchers found that one FIKK kinase specifically targets the amino acid tyrosine - something not previously observed in parasites. They believe this kinase evolved to interact with tyrosine-based signaling pathways in host cells.
Using a combination of computational and experimental approaches, including AlphaFold 2 for protein structure prediction, the team found that the kinases’ specificity for different protein targets is linked to small variations in a flexible “loop region.”
Although these small differences enable each kinase to recognize distinct proteins, the researchers also identified recurring patterns in the loop regions that distinguish FIKK kinases from human kinases, highlighting a possible drug target.
Blocking all FIKK Kinases
In collaboration with GlaxoSmithKline, the researchers screened a library of molecules known to inhibit human kinases to identify compounds that could block FIKK kinases. They identified three potential candidates and found that two of them effectively inhibited most FIKK kinases in vitro.
The next step is to modify these compounds to make them suitable for use in humans. Targeting all FIKK kinases simultaneously may offer a promising therapeutic strategy to reduce malaria symptoms.
About 1 million years ago, Plasmodium crossed from birds into great apes. With this cross, the FIKK kinase family expanded to enable infection of our closest relatives. A relatively short time ago, Plasmodium falciparum crossed from great apes to humans, and we have shown that the kinases needed for survival in great apes are still required for survival in humans. This suggests that targeting shared properties of FIKK kinases could stop P. falciparum from remodelling the host cell.
Moritz Treeck, Gulbenkian Institute for Molecular Medicine
Hugo Belda, former postdoctoral research assistant at the Crick, now postdoctoral researcher at GIMM and co-first author with David Bradley, added, “This project has been very collaborative across multiple institutions and disciplines, resulting in a holistic investigation into P. falciparum evolution. Current drugs mostly target single proteins, which makes the emergence of drug resistance more likely. Developing compounds which target several proteins at once, like those blocking all FIKK kinases, may be one way to tackle this problem.”
Source:
Journal reference:
Belda, H., et al. (2025) The fast-evolving FIKK kinase family of Plasmodium falciparum can be inhibited by a single compound. Nature Microbiology. doi.org/10.1038/s41564-025-02017-4.