Although antibiotics are known to save lives, their use also supports the evolution and spread of antibiotic-resistant strains.
The Centers for Disease Control and Prevention report that every year, antibiotic-resistant bacteria affect around 2.8 million people in the United States, causing mortality of over 35,000.
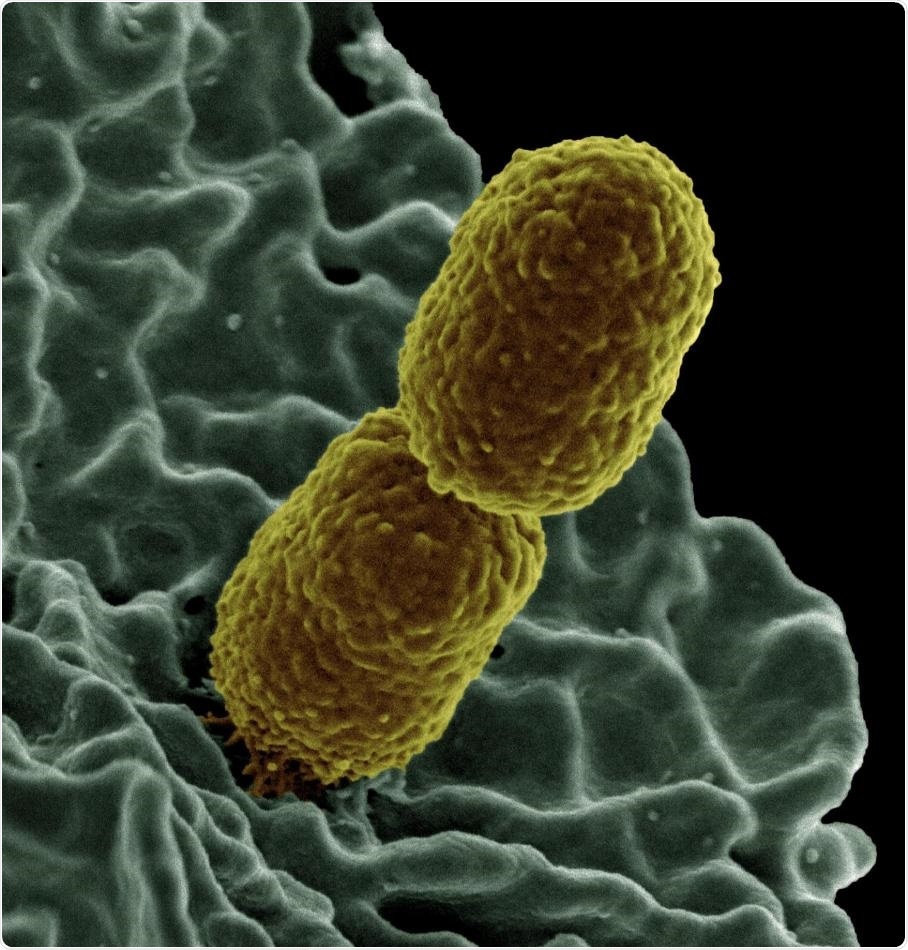
This is a colorized scanning electron micrograph image of Klebsiella pneumoniae interacting with a human neutrophil, a type of white blood cell. Image Credit: National Institute of Allergy and Infectious Diseases.
Infections caused by multidrug-resistant (MDR) bacteria, resistant to two or more antibiotics, are specifically challenging to treat.
Researchers from the University of Washington and the University of Idaho have identified how MDR bacteria readily evolve.
The researchers describe, in a paper published recently in Nature Ecology & Evolution, prolonged exposure of a bacterial pathogen, which was previously resistant to an antibiotic, to the same antibiotic enhances its ability to retain its resistance gene, while making the pathogen more readily absorb and maintain resistance to a second antibiotic and turn into an MDR strain.
The researchers conducted experiments that suggested that the long-time exposure to one specific type of antibiotic typically “primed” the bacteria.
Due to this priming effect, the bacteria acquire resistance to other antibiotics, even if they are not exposed to the antibiotic further. This enabled the strain to retain those antibiotic-resistance traits for generations.
Exposure to antibiotics appears to select indirectly for more stable antibiotic resistance systems. A more stable system in a strain will increase the chances that it will acquire resistance to multiple antibiotics.”
Benjamin Kerr, Study Co-Senior Author and Professor of Biology, University of Washington
The study outcomes reveal the effect of antibiotic exposure on the evolutionary dynamics of bacteria.
This could help explain not only the rise of multidrug resistance in bacteria, but also how antibiotic resistance persists and spreads in the environment—in health care settings, in soil from agricultural runoff—even long after the antibiotic exposure has ended.”
Eva Top, Study Co-Senior Author and Professor of Biology, University of Idaho
The researchers investigated a common mechanism for the distribution of antibiotic resistance—plasmids. Plasmids are circular strands of DNA that can include several types of genes, such as genes for antibiotic resistance. Bacteria tend to exchange plasmids easily within species.
However, plasmids have their own disadvantages, and earlier studies have revealed that bacteria lose them readily.
Even though they can carry beneficial genes, plasmids can also interfere with many types of processes inside a bacterial cell, such as metabolism or DNA replication. So, scientists have generally thought of plasmids as costly and burdensome to the host cell.”
Hannah Jordt, Study Lead Author and Research Scientist in Biology, University of Washington
Researchers from the University of Washington and the University of Idaho analyzed Klebsiella pneumoniae cells with a chloramphenicol-resistance plasmid and E. coli cells with a tetracycline-resistance plasmid.
Both the hosts had not been developed in the presence of antibiotics earlier and did not exhibit any loyalty to their plasmids. Following nine days of exposure to an antibiotic-free environment, the fragment of Klebsiella that still has a plasmid reduced to less than 50%. In the case of E. coli, it was found to be less than 20%.
Upon exposing the strains to antibiotics and growing each of them for 400 generations in their respective antibiotic, the strains exhibited higher affinity toward their plasmids even when they were no more exposed to the antibiotic.
When they were held in an antibiotic-free growth medium for nine days, over half of the Klebsiella and E. coli cells retained their respective plasmid.
According to Jordt, “Of course, the cells needed their plasmids to help them survive the antibiotic exposure. But even after we took away that selective pressure, both strains retained their plasmids at significantly higher levels than they had before the antibiotic exposure.”
Other experiments that were conducted demonstrated that exposure to antibiotics increases the occurrence of MDR Klebsiella. Even in the absence of exposure to antibiotics, Klebsiella pneumoniae gained multiple plasmids.
For instance, when antibiotic-naive E. coli and Klebsiella plasmid bearing strains were grown together, a small portion of Klebsiella turned MDR by holding onto their chloramphenicol-resistance plasmid and acquired tetracyclin-resistance plasmids from E. coli.
The experiment was repeated using bacteria exposed to antibiotics, finding nearly 1,000 times more MDR Klebsiella.
Prior long-time exposure to just one antibiotic known as chloramphenicol increased the chances that chloramphenicol-resistance Klebsiella would acquire the tetracycline-resistance plasmid from E. coli in the absence of antibiotics.
The experiments also showed that when MDR cells were grown in the absence of antibiotics, Klebsiella exposed to chloramphenicol had more affinity to both resistance plasmids.
According to the researchers, evolution can account for both the persistence of the antibiotic-resistance plasmids and the increase of MDR in Klebsiella. They added that this exposed the strains to their respective antibiotic chosen for mutations in their genomes to reduce the conflict between host and plasmid, which was less problematic to keep that plasmid and also others.
Kerr stated, “We believe that by stabilizing one plasmid, these mutations make them more likely to stabilize additionally acquired plasmids.”
Further experiments may help identify the particular mutations that assisted Klebsiella and E. coli to retain their plasmids, and the reason behind Klebsiella being more able to evolve into an MDR strain than E. coli.
Although exposure to antibiotics created selective pressure to hold the plasmids and acquire new ones, there was still an extensive variation in the way different species retain, share, and acquire plasmids.
Top added, “There are many, many details to be worked out in the future. But what we see here is that even short-term exposure to just one antibiotic accelerates the development of multidrug resistance, which should give us pause as we use these drugs in health care, agriculture, and other settings.”
Source:
Journal reference:
Jordt, H., et al. (2020) Coevolution of host–plasmid pairs facilitates the emergence of novel multidrug resistance. Nature Ecology & Evolution. doi.org/10.1038/s41559-020-1170-1.