For a long time, scientists have largely been producing transcriptomic data (that is, the messenger RNA (mRNA) converted into a protein) from cancer tissues, in an attempt to find clues about modified cellular pathways that may be fueling cancer behavior and can perhaps represent new therapeutic targets.
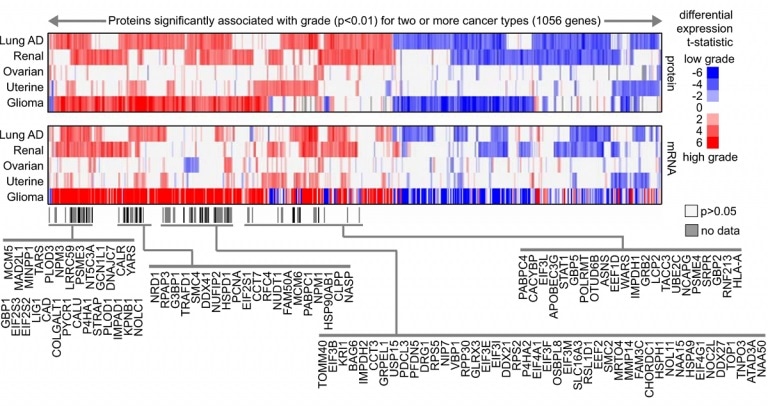
Proteins shared among the cancer type-specific grade comparing higher grade versus lower grade (red, higher expression with higher grade; white, not significant with p > 0.05.). Image Credit: Monsivais, et al/Oncogene.
While this method has been quite valuable, scientists from Baylor College of Medicine believe that transcriptomic data alone may not disclose the entire story behind cancer.
There are two notable aspects of this study. One is that we explored the proteomic landscape of cancer looking for proteins that were expressed in association with aggressive forms of cancer.”
Dr Chad Creighton, Professor of Medicine and Co-Director, Cancer Bioinformatics, Dan L Duncan Comprehensive Cancer Center, Baylor College of Medicine
Dr. Creighton, who is also the co-corresponding author of the study, added, “We analyzed protein data that included tens of thousands of proteins from about 800 tumors including seven different cancer types—breast, colon, lung, renal, ovarian, uterine, and pediatric glioma - made available by the Clinical Proteomic Tumor Analysis Consortium (CPTAC) mass-spectrometry-based proteomics datasets.”
Computational analysis of the CPTAC dataset helped detect proteomic signatures linked with severe types of cancer. Such signatures indicate modified cell pathways that may be fueling aggressive cancer behavior and could represent new therapeutic targets. Each type of cancer had a distinct proteomic signature for its aggressive form. Fascinatingly, some of the signatures were common to different forms of cancer.
The other feature of this research work was to provide evidence that proteomic analysis was a valuable method to detect the drivers of aggressive diseases that could possibly be exploited to regulate the growth of cancer.
That's exactly what we were able to do with this new, very powerful dataset. We focused on the uterine cancer data for which the computational analysis identified alterations in a number of proteins that were associated with aggressive cancer. We selected protein kinases, enzymes that would represent stronger candidates for therapeutics.”
Dr Diana Monsivais, Study Co-Corresponding Author and Assistant Professor of Pathology and Immunology, Baylor College of Medicine
Among the hundreds of preliminary candidates, four kinases were chosen for functional studies in uterine cancer cell lines. The researchers observed that kinases not only were expressed in uterine cancer cells lines, but that exploiting the expression of certain kinases also decreased the ability or survival some uterine cancer cells to migrate. Cell migration is a feature of cancer cells that helps them to proliferate to other tissues.
This study is the outcome of a productive partnership between two centers at Baylor College of Medicine—the Dan L Duncan Comprehensive Cancer Center and the Center for Drug Discovery.
Chad conducted this wonderful analysis on new CPTAC datasets and was interested in validating it in a human cancer. He approached us about performing functional studies to determine whether some proteins could translate to new targets for endometrial cancer. Our experiments provided proof-of-concept that proteomics analysis is a useful strategy not only to better understand what drives cancer, but to identify new ways to control it or eliminate it.”
Dr Diana Monsivais, Study Co-Corresponding Author and Assistant Professor of Pathology and Immunology, Baylor College of Medicine
“Historically, researchers have only been generating transcriptomic data (the messenger RNA (mRNA) that is translated into protein). Looking at the protein data itself, which is made available by the CPTAC, enables researchers to extract a new layer of information from these cancers,” added Creighton.
“In this study, we compared mRNA and protein signatures and, although in many cases they overlapped, about half the proteins in the proteomic signatures were not included in the corresponding mRNA signature, suggesting the need to include both mRNA and protein data in cancer studies,” Creighton concluded.
Source:
Journal reference:
Monsivais, D., et al. (2021) Mass-spectrometry-based proteomic correlates of grade and stage reveal pathways and kinases associated with aggressive human cancers. Oncogene. doi.org/10.1038/s41388-021-01681-0.