Bitter, sweet, sour, salty, and umami comprise the five tastes humans can recognize. But the way these tastes and flavors are experienced will be unique to the individual.
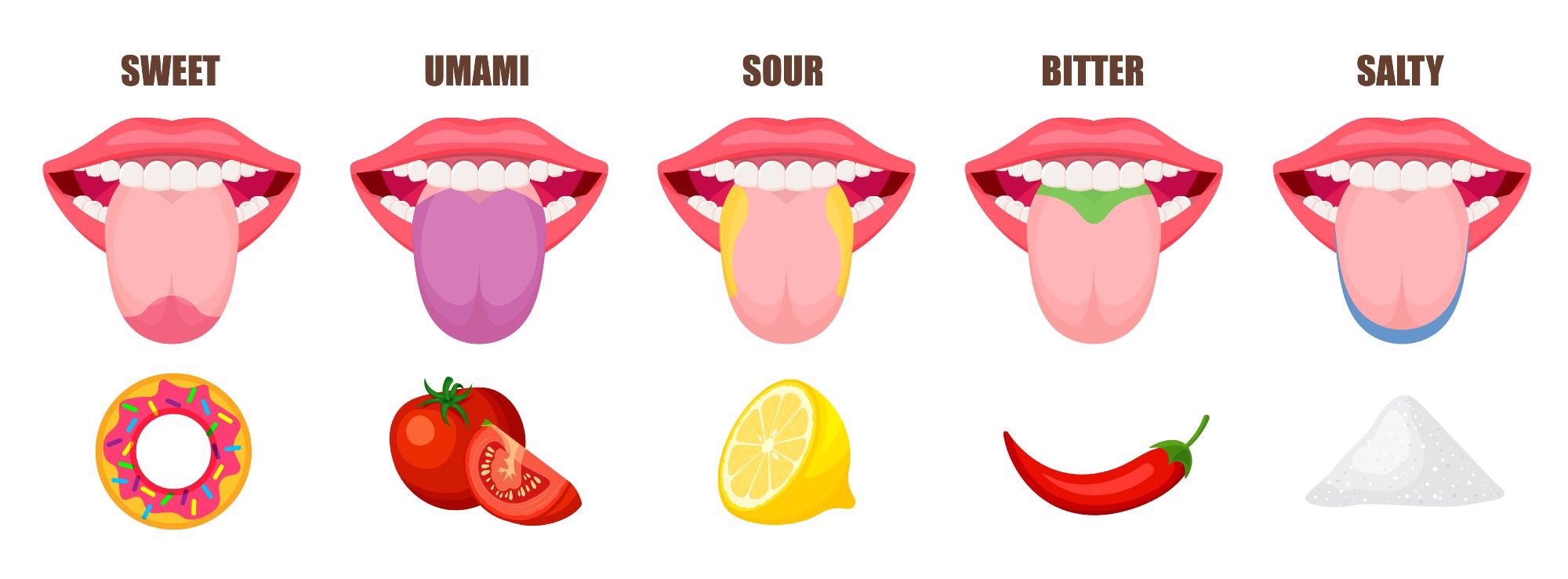
Image Credit: Yuliia Konakhovska/Shutterstock.com
Differences in the perception of taste will influence food habits and preferences. A perhaps obvious example of this is cilantro/coriander. For some people, cilantro has a distinct soapy taste. Genetic variation in the region of olfactory-receptors has been associated with the soapy flavor that some experience.
Comprising over 400 known functional alleles, olfactory receptors can impart enormous variation on taste and smell. Similarly, variation in the TAS1R and TAS2R family of receptors influence the perception of bitter, sweet, and umami. Recent evidence also alludes to the role of taste and smell receptors when expressed outside the gustatory system.
The TAS2R family of G-protein coupled receptors (GPCR)
Taste begins with transmitting information by ion channels or GPCRs; of which TAS1R and TAS2R are both the latter. Binding extracellular stimuli, the GPCRs transduce signaling by conformational changes. Structural changes in the receptor activate it, allowing for intracellular G-proteins to bind.
Acting as a guanine nucleotide exchange factor, the receptor induces conformational changes in the G-proteins to promote the release of GDP, and binding of GTP. GTP binding triggers activation of the G proteins and dissociation of the α subunit. The activated α and βγ subunits go on to regulate various downstream signaling cascades.
Genetic variation in TAS2R
In TAS2R there are 25 identified alleles associated with the perception of bitter taste; as well as G protein components α-gustducin, β3, and γ13. Polymorphisms in these genes are linked to bitter taste modality. A classic example of this is the ability or inability to taste phenylthiocarbamide (PTC). With two main alleles termed the "taster" and "non-taster", genetic variation in the TAS2R38 influences response to bitter compounds PTC, as well as 6-n-propylthiouracil (PROP).
A single nucleotide polymorphism (SNP) on TAS2R38 results in amino acid substitution, transcribing alanine, valine, and isoleucine in place of proline, alanine, and valine. The resulting non-functional variant confers an inability to perceive the bitterness of PTC and PROP.
TAS2R variant and food preferences
In combination with the salivary trophic factor, gustin, TAS2R38 variants can greatly influence an individual’s sensitivity to bitter taste. One proposed mechanism of how these genes influence taste is through the morphology of papillae. These raised lingual structures contain taste buds; in those with have a high sensitivity to PROP, the density of the papillae is greater.
The TAS2R38 variant, is perhaps the most widely studied, but variation in any of the 25 known TAS2R genes can affect the perception of bitterness. Functional variants of TAS2R43 have been linked to the perception of bitterness in coffee, most likely through caffeine. SNPs in this gene, particularly the H212R variant, have been associated with reduced in vitro protein activity; this non-functional H212R variant has been linked to disliking coffee.

Image Credit: KK_face/Shutterstock.com
Bitter taste receptors in the stomach and gut
Expressed in the mouth and stomach, TAS2R43 has been linked to caffeine-induced gastric acid secretion. However, this secretion is site-dependent, with caffeine in the mouth signaling inhibition of secretion. When the bitter compounds reach the stomach, activated TAS2R43 increases the secretion of gastric acid. In this way, activating the TAS2R43 receptor could have therapeutic potential in regulating gastric pH.
TAS2Rs found in the gut, are known to influence metabolism. Expression of TAS2R4, TAS2R10, and TAS2R38 has been identified in the gut; with tissue-dependent, altered expression of these in obese non-diabetic patients. Activation of TAS2R5 and TAS2R10 in fundic cells stimulates ghrelin secretion, a “hunger hormone" responsible for stimulating appetite, increasing food intake, and promoting fat storage.
Clinical applications of TAS2R
Activated TAS1R and TAS2R in the gut have been identified as promoting secretion of insulinotropic hormones, or incretins. For individuals with type 2 diabetes, this incretin pathway offers new potential therapeutic targets. Though no drugs have yet been developed to exploit this pathway, activation of TAS2Rs in the gut of diet-induced obese mice has shown promising results.
Activating TAS2Rs with bitter stimuli in the gut is seen to elevate intracellular calcium and stimulate the secretion of glucagon-like peptide 1 (GLP-1). By altering enteroendocrine hormone release and bile acid metabolism, TAS2Rs are thought to ameliorate the symptoms of diabetes. Following acute treatment, mouse models displayed increased glucose-dependent insulin secretion and improved glucose tolerance. Chronic treatment stimulated sustained release of GLP-1, with mice losing weight and fat mass; as well as having increased energy expenditure, glucose tolerance, and insulin sensitivity.
Outside of the gustatory system, TAS1Rs and TAS2Rs are thought to perform many functions. Variations in TAS2R genes have been identified as important predicting factors for the course of chronic rhinosinusitis. TAS2Rs are also thought to play an important role in cancer, with their presence in breast cancer cells, ovarian cancer cells, and neuroblastoma, providing novel therapeutic targets or oncological markers.
Sources:
- Jeruzal-Świątecka, J., et al. (2020) Clinical Role of Extraoral Bitter Taste Receptors. Int J Mol Sci. doi:10.3390/ijms21145156
- Dotson, C.D., et al. (2010) T1R and T2R receptors: the modulation of incretin hormones and potential targets for the treatment of type 2 diabetes mellitus. Curr Opin Investig Drugs. 11(4), pp. 447-454.
- Kok, B.P., et al. (2018) Intestinal bitter taste receptor activation alters hormone secretion and imparts metabolic benefits. Molecular Metabolism doi.10.1016/j.molmet.2018.07.013.
- Liszt, K.I., et al. (2017) Caffeine induces gastric acid secretion via bitter taste signaling in gastric parietal cells. Proc Natl Acad Sci USA. doi:10.1073/pnas.1703728114.
- Judit, D., et al. (2019) Genetic Background of Taste Perception, Taste Preferences, and Its Nutritional Implications: A Systematic Review. Frontiers in Genetics. doi:10.3389/fgene.2019.01272.
- Melis, M., et al. (2013) The Gustin (CA6) Gene Polymorphism, rs2274333 (A/G), as a Mechanistic Link between PROP Tasting and Fungiform Taste Papilla Density and Maintenance. PLOS ONE. doi:10.1371/journal.pone.0074151.
- Pirastu, N., et al. (2014) Association analysis of bitter receptor genes in five isolated populations identifies a significant correlation between TAS2R43 variants and coffee liking. PLoS One. doi:10.1371/journal.pone.0092065.
- Eriksson, N., et al. (2012) A genetic variant near olfactory receptor genes influences cilantro preference. Flavour. doi: 10.1186/2044-7248-1-22.
- Melis, M., et al. (2019) TAS2R38 bitter taste receptor and attainment of exceptional longevity. Sci Rep. doi:10.1038/s41598-019-54604-1.
- Khan, A.M., et al. (2019) Impact of Fungiform Papillae Count on Taste Perception and Different Methods of Taste Assessment and their Clinical Applications: A comprehensive review. Sultan Qaboos Univ Med J. doi:10.18295/squmj.2019.19.03.003.
Further Reading
Last Updated: Feb 24, 2022