Multiple disease detection and classification via microbiome search. Image Credit: SU Xiaoquan.
A microbiome search-based method has now been proposed by an international team of scientists to analyze the abundance of available health data to identify and diagnose human diseases. This method involves the use of the Microbiome Search Engine (MSE; http://mse.ac.cn).
Microbiome-based disease classification depends on well-validated, disease-specific models or markers. However, current models are lacking that information for many diseases.”
Dr SU Xiaoquan, Single-Cell Center, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences
SU added that multiple diseases could share similar biomarkers, which are the microorganisms that signify that something is unusual, for example, a mutated protein identified in cancer cells. Thus, researchers find it difficult to accurately classify each one.
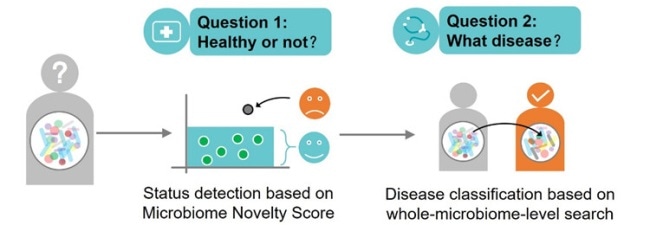
SU and his collaborative software team from Single-Cell Center, QIBEBT and Center for Microbiome Innovation (CMI), University of California at San Diego (UCSD) overcame the issues for disease detection and classification by developing a new search method based on the whole microbial community within a human body, called the microbiome. Every individual has a microbiome, although they do not have a disease.
In conventional models, samples from healthy subjects are compared with individuals affected by particular diseases. The scientists can determine the microbiome state related to the disease across different cohorts or sequencing platforms by using this new approach of searching based on the particular outlier rather than known biomarkers that code for various diseases.
As part of the new method, the researchers use a two-step procedure to diagnose disease. Firstly, a baseline database of healthy individuals is searched to identify any particular microbiome that is an exception—or any familiar anomaly that differentiates the microbiome from a healthy state.
Secondly, they search for that exception in a database of disease-specific examples.
Our strategy’s precision, sensitivity and speed outperform model-based approaches.”
Dr SU Xiaoquan, Single-Cell Center, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences
The outcomes of this search can offer quick estimates to help clinicians in the diagnosis and treatment of diseases.
This search-based strategy shows promise as an important first step in microbiome big data-based diagnosis In light of the general shift of microbiome-sequencing focus from healthy to diseased hosts, the findings here advocate for adding more baseline samples from across different geographic locations.”
Rob Knight, Director, Center for Microbiome Innovation, University of California at San Diego
This was agreed upon by XU Jian, Director of Single-Cell Center, QIBEBT. The next step of the joint Sino-USA team is to encourage their collaborators to take a combined effort to continue the expansion of the microbiome database, to include every ecosystem and every population on the planet.
According to XU, “With Microbiome Search Engine, performing a search can become as standard and enabling for new microbiome studies as performing a BLAST against your new DNA sequence is today.”
This study was financially supported by the National Natural Science Foundation of China, the Chinese Academy of Sciences, the National Institutes of Health, the National Science Foundation, the Alfred P. Sloan Foundation, and the National Health and Medical Research Council.
Source:
Journal reference:
Su, X., et al. (2020) Multiple-Disease Detection and Classification across Cohorts via Microbiome Search. mSystems. doi.org/10.1128/mSystems.00150-20.