The 3D structure of DNA is typically established by a sequence of spacial rules on the basis of specific protein sequences and their order.
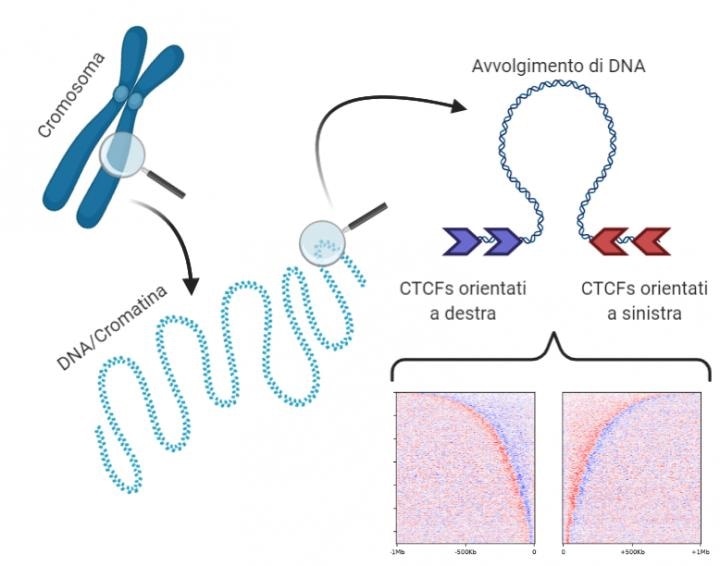
CTCF proteins isolate the various topological DNA domains. The study found that topological domains can be divided into two sections with specular grammatical sequences, delimited by two “barriers” and with a “reversal point” in the middle separating the right (blue) and left (red) CTCF sequences. The human genome compresses following a “grammar” logic comprising CTCF sequences, orientation, and the distance between them. Image Credit: Luca Nanni.
This was the result of a study recently published in the Genome Biology journal by Luca Nanni, a PhD student in Computer Science and Engineering at The Polytechnic University of Milan (Politecnico di Milano), along with Professors Stefano Ceri from the same University and Colin Logie from the University of Nijmegen.
Our study’s greatest innovation lies in having identified precise rules for the disposition of CTCF proteins. The beauty and simplicity of CTCF’s grammar shows us how nature and evolution produce regularity and incredibly ingenious and functional systems. Knowing these rules allow CTCF sequences to be engineered to obtain the desired DNA three-dimensional structure.”
Luca Nanni, Study First Author and PhD Student, Department of Computer Science and Engineering, The Polytechnic University of Milan
Nanni continued, “For example, it should be possible to make two disconnected genes interact. Moulding DNA structure will open doors to the creation of pharmaceuticals for the treatment of diseases such as cancer.”
The DNA molecule, which would measure 2 m in length if fully unfolded, wraps itself on the basis of a complex system that preserves its correct reading and accessibility to remain in the nucleus of the cell.
Topological domains are significant in the analysis of the 3D genome structure. These domains are known to aggregate DNA zones that have analogous behavior and roles. For instance, genes that have analogous functions may probably reside in the same topological domain.
We focused on some specific DNA sequences that encode for the CTCF protein. This protein isolates portions of DNA creating barriers between the various topological domains. With the help of computer simulations and the creation of a model for classifying these proteins according to their orientation, we identified a surprising regularity in their arrangement along the DNA sequence.”
Luca Nanni, Study First Author and PhD Student, Department of Computer Science and Engineering, Polytechnic University of Milan
The research work demonstrated that topological domains could be reconstructed based on the order and orientation of these DNA sequences. The human genome condenses following a “grammar” logic that contains CTCF sequences, the distance between them, and orientation.
Source:
Journal reference:
Nanni, L., et al. (2020) Spatial patterns of CTCF sites define the anatomy of TADs and their boundaries. Genome Biology. doi.org/10.1186/s13059-020-02108-x.