A novel genetic analysis of blood protein concentrations, headed by scientists in the MRC Integrative Epidemiology Unit (MRC-IEU) at the University of Bristol, has shown how genetic information can be applied to support the prioritization of drug targets by detecting the causal impacts of proteins on various diseases.
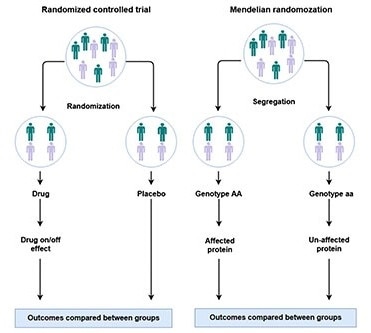
Comparison between randomized controlled trial and the genetic approach “Mendelian randomization.” Image Credit: University of Bristol.
Bristol scientists, working in association with pharmaceutical firms, have created a detailed analysis pipeline by using the genetic prediction of protein concentrations for drug target prioritization, and have measured the ability of this method to decrease the failure rate of the development of drugs.
Genetic analyses of proteins are still in the nascent stage. The objective of this study, published in the Nature Genetics journal, was to determine whether genetic prediction of protein target effects could be used to predict the success rate of a drug trial.
Dr Jie Zheng, Professor Tom Gaunt, and collaborators from the University of Bristol teamed up with pharmaceutical firms to establish a multi-disciplinary partnership to deal with this scientific question.
The team used a set of genetic epidemiology methods, such as Mendelian randomization genetic colocalization, and constructed a causal network of 1002 plasma proteins on a total of 225 human diseases. While doing so, they detected 111 putatively causal impacts of 65 proteins on 52 diseases, spanning a broad range of disease areas. The study results can be accessed through EpiGraphDB.
According to the study’s lead author, Dr Zheng, their predicted causal effects of proteins on human diseases could be utilized to estimate the effects of drugs that target these proteins.
This analysis pipeline could be used to validate both efficacy and potential adverse effects of novel drug targets, as well as provide evidence to repurpose existing drugs to other indications. This study lays a solid methodological foundation for future genetic studies of omics.”
Dr Jie Zheng, Study Lead Author, University of Bristol
Dr Zheng continued, “The next step is for the analytical protocol to be used in early drug target validation pipeline by the study’s pharmaceutical collaborators. We hope that these findings will support further drug development to increase the success rate of drug trials, reduce drug cost and benefit patients.”
Our study used publicly available data published by many researchers around the world (collated by the MRC-IEU OpenGWAS database), and really demonstrates the potential of open data sharing in enabling novel discoveries in health research. We have demonstrated that this re-use of existing data offers an efficient approach to reducing drug development costs with anticipated benefits for health and society.”
Tom Gaunt, Professor of Health and Biomedical Informatics, University of Bristol
Professor Gaunt is also a member of the NIHR Bristol Biomedical Research Centre.
Source:
Journal reference:
Zheng, J., et al. (2020) Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nature Genetics. doi.org/10.1038/s41588-020-0682-6.