Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been shown to enter cells by interaction with angiotensin-converting enzyme 2 (ACE2), which is found on the surface of cells in the lungs and throughout the body.
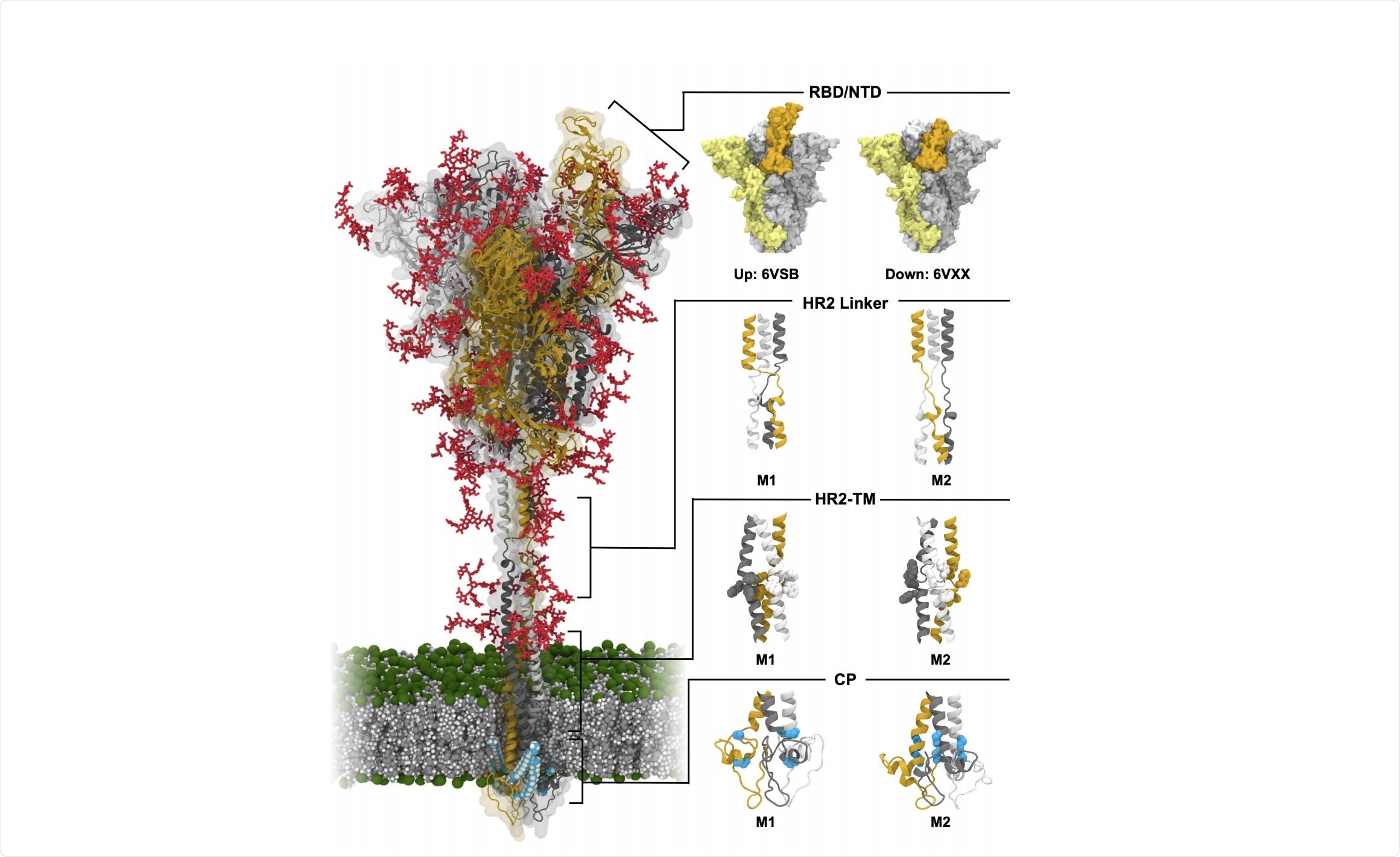
Image Credit: https://www.biorxiv.org/content/10.1101/2020.10.18.343715v1.full.pdf

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
The spike protein of SARS-CoV-2 is a class 1 fusion protein, constructed bearing two independent joints that allow flexibility, and coated with a variety of glycans that stabilize the open or closed states of the receptor-binding domain. It is a homo-trimer, with each monomer having two subunits and a binding site for host proteases.
The first subunit is responsible for membrane binding, bearing the signal peptide, N-terminal domain, and receptor-binding domain. The second subunit is responsible for membrane fusion, consisting of the fusion peptide, two heptad repeats, transmembrane domain, and cytoplasmic domain. When in a closed position the binding domains are occluded by adjacent binding domains, though once opened they no longer sterically hinder their neighbors and are conformationally variable.
Thus far, techniques such as cryogenic electron microscopy have largely elucidated the structure of the spike protein, though the fine details of flexible protein regions remain unresolved. Choi et al. (2020) utilized molecular dynamics simulations to provide insight into the molecular interactions taking place around the spike protein, specifically in the conformational variability of the protein stalk that allows the head orientational freedom and in the significance of glycan shielding towards antibodies. The group simulated spike proteins both in glycosylated and non-glycosylated forms, in open and closed states, for several 1.25 µs long simulations.
A pre-print version of the paper is available on the server bioRxiv*, while the article undergoes peer review.
What is the role of stalk flexibility?
16 conformational variations were selected to be as accurate to experimental data gathered from other sources as possible, representing each of the three binding domains being open or closed in differing configurations and with alternate structures of the cytoplasmic, transmembrane, and receptor binding domains.
Each variation underwent 1.25 µs long simulations, and the angle of the spike protein stalk and head monitored throughout. Interestingly, the head was shown to be largely stable and immobile throughout, changing the relative angle by less than a nanometer.
The stalk, however, was found to be highly flexible, in particular around the heptad repeats nearest the transmembrane domain (HR-TM) and below the head (HR-linker), creating a two-jointed bending and twisting system. Differing configurations of the heptad repeats were examined, a total of 27,000 configurations and the average angle between the membrane and stalk determined to be 48⁰, with the possibility of a full 90⁰ tilt.
Structural alignment studies between the stalk protein and ACE2 were subsequently performed, finding that the most probable angle to allow interaction was 46⁰. Spike proteins are generously spaced out across the viral surface, having only one for every 1000 nm2 of surface area. This work suggests that such ample space encourages the adoption of the angle of best interaction with ACE2, contributing to the high infection rate observed in SARS-CoV-2.
What is the influence of glycans?
Glycans on the surface of the spike protein were found to contribute to the motion of the receptor binding and N-terminal domains. In comparing models of the spike protein in its glycosylated and non-glycosylated form it was observed that the receptor-binding domain of those without glycans preferentially adopted a closed conformation, indicating that the open form is unstable without the presence of glycans.
Three N-glycans in particular were identified as having pivotal roles in the conformational change of the receptor-binding domain: N165, N234, and N343. N165 and N234 glycans are located below the open-state receptor binding domain, contributing towards holding it open, while in the closed state the receptor-binding domain forms a sandwich arrangement with these glycans, holding it closed.
N343 glycan is oriented outwards from the domain on which it is located, forcing its interactions to be with neighboring domains. When the native domain is open the glycan cannot interact, though when closed may interact with an open or closed neighboring domain to maintain this state, contributing towards keeping it open or closed, respectively.
Since it is observed that the binding domains are constantly transitioning between open and closed states the authors suggest that these glycans serve to act as a clutch, temporarily holding a particular state and thereby modulating the amount of time spent in either state.
Other glycans were identified as having significant and specific roles in shielding the spike protein. Four receptor binding domain targeting antibodies were selected for applicability and similarity to ACE2 and superimposed over the receptor-binding domains to determine likely glycan-antibody clashes. Some were found to play a clear shielding role in certain situations, for example, N165 glycan is likely to interact with the antibodies only when the receptor-binding domain is closed, while N122 glycan only encounters severe clashes when the domain is open.
The authors make a strong note, however, that accessible surface area calculations often over-attribute the effect of glycan shielding on antibody binding, while this work suggests that minor clashes are insufficient to entirely block antibody binding since glycans are highly flexible and do not represent a severe hurdle. Furthermore, antibody-glycan interactions actually contribute to antibody binding in some situations.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
https://www.biorxiv.org/content/10.1101/2020.10.18.343715v1