The capacity of the COVID-19 virus’s spikes to bind to human cells may be hampered by molecules from the same family as the anticoagulant medication heparin. This might be used to treat patients who are suffering from the virus’s severe symptoms, as well as any new strains.
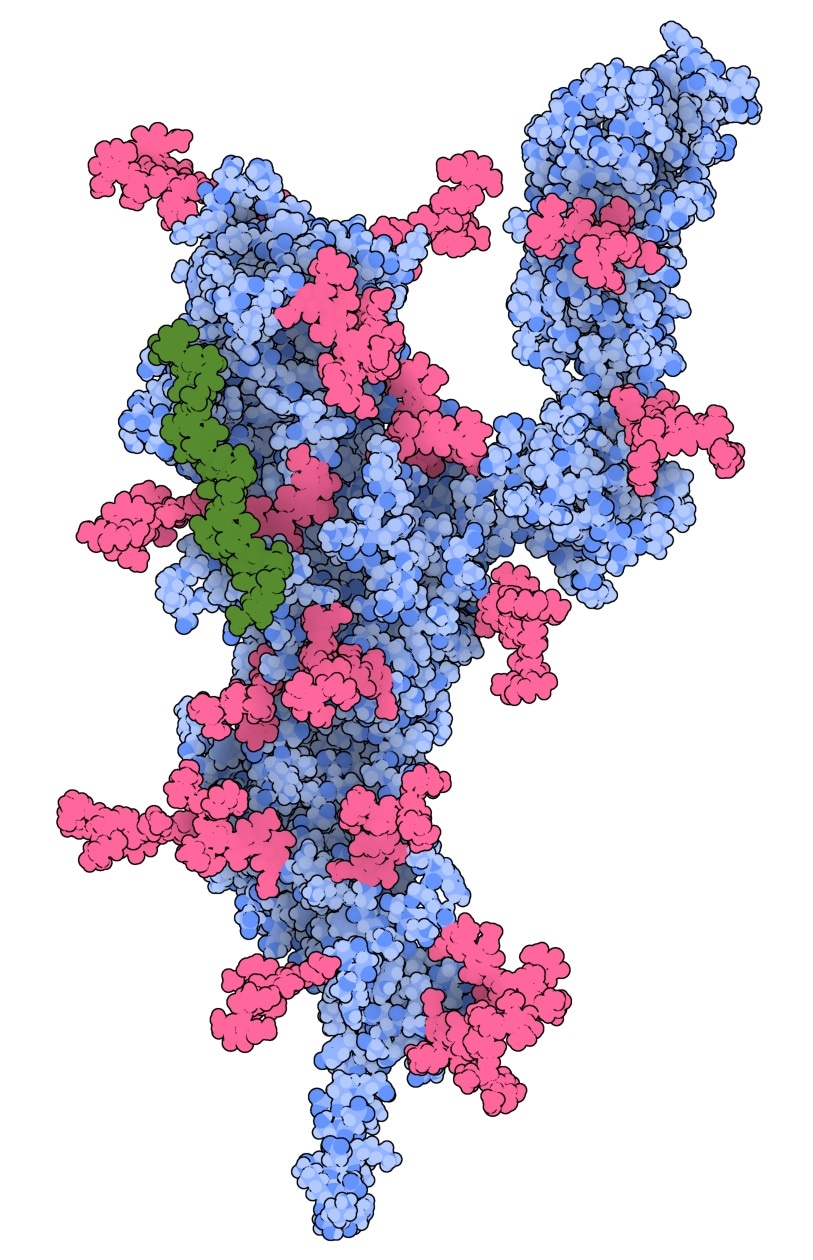
3D model showing heparan sulphate (green) binding to the N-terminal receptor-binding domain of the SARS-CoV-2 spike protein. Image created using 3D protein imager app The Protein Imager. Image Credit: Queensland University of Technology.
The study team discovered a new binding site on the SARS-CoV-2 spike protein, according to QUT PhD researcher Zachariah Schuurs.
Binding of the CoV-2 spike protein to heparan sulphate (HS) on cell surfaces is generally the first step in a cascade of interactions the virus needs to initiate an infection and enter the cell. Most research has focused on understanding how HS interacts on the receptor-binding domain (RBD) and furin cleavage site of the SARS-CoV-2 virus’s spike protein, as these typically bind different types of drugs, vaccines and antibodies.”
Zachariah Schuurs, PhD Researcher, Queensland University of Technology
“We have identified a novel binding site on the N-terminal domain (NTD), a different area of the virus’s spike that facilitates the binding of HS. This helps to better understand how the virus infects cells. The NTD is also a part of the spike protein that frequently mutates,” added Schuurs.
Schuurs continued, “Some antibodies in the blood that neutralize the viruses bind to the same region of the NTD. Therefore, targeting the NTD site with molecules like heparin (or heparin mimetics), a known anti-coagulant drug similar to HS, is a possible strategy to stop the virus binding to cells and infecting them.”
COVID-19 vaccines, despite their worldwide success, are still far from being publicly available, according to Dr Neha Gandhi of the QUT Centre for Genomics and Personalised Health.
We need alternative antiviral strategies to prevent the spread of COVID-19 and treat infected people. Epidemiologists believe that persistent low-vaccine coverage in many countries will make it more likely for vaccine-resistant mutations to appear. Variants of concern have already emerged in South Africa, the US, India and Brazil. In this regard, alternate antiviral strategies are strongly needed to prevent the spread of COVID-19 and to treat people with COVID-19.”
Dr Neha Gandhi, Centre for Genomics and Personalised Health, Queensland University of Technology
“Most SARS-CoV-2 variants have acquired a positively charged mutation in the spike protein. Molecules like heparin and its mimetics are negatively charged and therefore, these molecules could be used to treat people with severe effects of the virus and any emerging variants,” Dr Gandhi explained.
“Our research indicates that molecules that mimic the 3D structure of heparin with different sulphur chemistry, might be potential broad-spectrum antiviral drugs for COVID-19 and other emerging viral threats via direct interaction with the virus itself,” added Dr Gandhi.
“Evidence of a putative glycosaminoglycan binding site on the glycosylated SARS-CoV-2 spike protein N-terminal domain,” a multi-national study published in the Computational and Structural Biotechnology Journal, used both computational and experimental methodologies to corroborate the findings.
The researchers employed supercomputers from NCI Gadi and QUT Lyra to simulate how HS and its inhibitors, such as heparin, interact with the spike protein.
The project was conceptualized by researchers from the University of Queensland (Glycochemistry), Curtin University (Immunology), Zucero Pharmaceuticals, and QUT. The QUT research team used computational molecular simulations to test and confirm the notion that heparan sulphate could establish a bridge between the newly discovered binding site and another critical location on the spike in the virus’s infection.
Bioinformatics support was provided by collaborators from the National Institute for Biological Standards and Control in the United Kingdom and the Istituto di Ricerche Chimiche e Biochimiche “G.Ronzoni” in Italy.
Laboratory investigations undertaken by collaborators at the University of Liverpool and Keele University in the United Kingdom verified the hypothesis.
Source:
Journal reference:
Schuurs, Z. P., et al. (2021) Evidence of a putative glycosaminoglycan binding site on the glycosylated SARS-CoV-2 spike protein N-terminal domain. Computational and Structural Biotechnology Journal. doi.org/10.1016/j.csbj.2021.05.002.