Being able to analyze gene expression patterns is essential for understanding protein function, biological pathways, and cellular responses to external and internal stimuli. This article aims to provide a brief overview of the processes that underpin gene expression and the techniques that can be used to quantify the expression of specific genes.
Gene Expression. Image Credit: udaix/Shutterstock.com
What is gene expression?
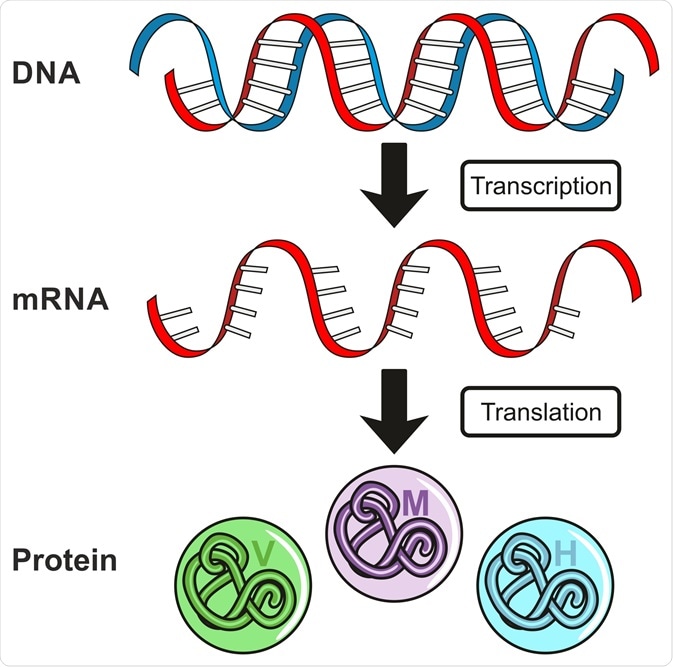
Gene expression controls the amount and type of proteins that are expressed in a cell at any given point in time. This is in turn controlled by regulatory mechanisms that control the synthesis and degradation of proteins within a pathway.
The process of gene regulation includes 1) transcription, the conversion of DNA to RNA, and 2) translation, the conversion of RNA to proteins. Aside from gene expression, protein levels can also be dictated by the amount of RNA in a cell.
DNA to RNA: Transcription
DNA transcription was initially observed using the method of electron microscopy in 1970. The resolution of these early microscopes was low, and DNA appeared as "trunks" with extended branches of nucleic acids. The addition of DNAses degraded the trunks, while RNAases removed the branches.
Although DNA molecules are double-stranded, only one strand acts as a template for the process of transcription. This strand is referred to as the “template strand”. The “nontemplate” strand is called the coding strand, as the sequence of this strand is the same as the sequence of the RNA molecule that is generated. In many cases, the template strand for one gene can also be the non-coding strand for other genes that are present in the chromosome.
The process of transcription begins by the attachment of the RNA polymerase to the template DNA strand which leads to the generation of complementary RNA molecules. The RNA polymerases are large molecules that consist of almost a dozen subunits along with other factors when attached to the DNA strand.
The number of polymerases differs in prokaryotes and eukaryotes: bacteria (which are prokaryotes) have only one RNA polymerase, while eukaryotic cells have RNA Pol I, II, and III. RNA Pol I encodes 47S ribosomal RNA, RNA Pol II encodes messenger RNAs, and RNA Pol III encodes for the 5S ribosomal RNA and transfer RNA.
Initiation, Elongation, and Termination
First, the RNA polymerase binds to a region present upstream to the actual coding sequence. This region is called a promoter. For binding to the DNA, RNA Pol binds to the “sigma” subunit forming a holoenzyme that can unwind the double helix of the DNA.
The unwinding is necessary to get access to the gene, and the sigma factor ensures that the RNA Pol binds to the correct region in the DNA. As the transcription proceeds, the helix unwinds, RNA Pol reads the template and adds the nucleotides on the 3’ end.
An average of 42-54 nucleotides is added per second when the temperature is 37 °C. This step (known as elongation) coordinates multiple events, some of which prevent errors during this process.
Termination can be of two types: in Rho-independent termination, the presence of inverted repeat sequences causes the transcribed RNA sequences to fold on themselves forming hairpin loops. This causes the RNA pol to detach, leading to termination. In the case of Rho-dependent termination, the rho factor releases the newly formed mRNA from the DNA by unwinding it.
RNA to Protein: Translation
Transcription produces a single molecule of mRNA, which can be defined as a single-stranded copy of a gene. mRNA then undergoes translation to produce a protein. Every three bases in the mRNA sequence constitute an amino acid – thus, the translation produces a string of amino acids.
The process of translation occurs in the ribosome. So, after the process of transcription which occurs in the nucleus, the mRNA travels outside the nucleus to the ribosome. In prokaryotes, as there is no separation or compartmentalization of the nucleus, the process of translation starts even as the DNA is being transcribed.
The ribosome consists of two subunits: small and large. The smaller subunit and the initiator transfer RNA (tRNA) assemble on the mRNA strand. The small subunit has an amino acid site (A), a polypeptide site (P), and an exit site (E). The aminoacyl-tRNA binds to the mRNA at the A site. At the P site, the amino acid is transferred from the tRNA to the polypeptide chain.
Finally, E or exit site is the position of empty tRNA before it is released into the cytoplasm. The three termination codons at the end of protein-coding mRNA sequences are UAA, UAG, and UGA. These signify termination as there are no tRNAs to recognize these codons.
Image Credit: Meletios Verras/Shutterstock.com
RNA Splicing: Multiple Proteins from a Single RNA Sequence
In the case of eukaryotic genes, the RNA that is initially made from the DNA template undergoes processing before a mature messenger RNA is created. This processing involves RNA splicing where certain sequences are “spliced” or “removed”.
These sequences are introns which are noncoding in nature. The final sequences that are left in the mRNA are coding sequences or exons. The introns are cleaved at sites called splice sites that are present at the 5' and 3' end of the introns.
The common RNA sequence includes nucleotide GU at the 5’ end and AG at the 3’ end. This sequence is very important as any change can inhibit the spicing process.
The splicing process is catalyzed by small ribonucleoproteins known as “snRNPs” (usually pronounced “snurps”) and occurs in cellular machines called spliceosomes. The concept of splicing makes genes more “modular” where new combinations of exons can generate new proteins without changing or disrupting the old genes.
Measuring and Quantifying Gene Expression
The identity and levels of expressed genes can be critical to understanding any biological process. Since at any given point of time only a small fraction of genes are expressed, it is important to assess the gene expression profile.
To obtain a quantitative assessment of changes in the levels of mRNA, a sufficient quantity of either total or messenger RNA, probes that are specific to the required sequences, necessary controls, as well as a sensitive detection method are needed. Currently, the two major methods to quantitatively detect mRNA levels include electrophoretic methods (such as Northern blot), DNA microarray, and quantitative PCR.
Northern blot
Northern blot is a commonly employed tool. Its advantage lies in the fact that the transcript size is obtained using gel electrophoresis. This provides a crude verification method of the probe accuracy and also identifies the splice variants present in the RNA sample. However, it is labor-intensive and a large number of steps enables more experimental errors to creep in.
DNA microarrays
The starting point of a DNA microarray involves creating an array of sequences that correspond to the genes that need to be probed. These oligonucleotides are synthesized chemically and multiple such probes can be designed for each gene. Now it is possible to robotically print cDNA probes on a glass slide. Using this method it is possible to obtain the entire profile of gene expression in a single experiment.
Real-time PCR
A powerful method that has emerged to measure the mRNA levels is kinetic or real-time PCR. In the method, accumulated PCR products are monitored at the end of every cycle by employing fluorescence.
The fluorescence is initially indistinguishable from the background; however, after a certain number of cycles, the fluorescence starts increasingly exponentially before reaching a plateau. This increase in fluorescence can be used to quantitatively determine levels of mRNA.
Further Reading
Last Updated: Jan 27, 2021