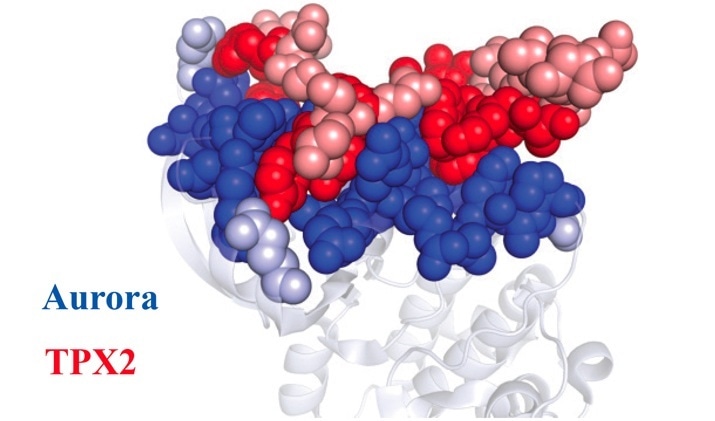
Once the Aurora A protein (blue) and TPX2 protein (red) learned how to fit together, they stayed that way forever. Dark dots represent amino acids that remain unchanged throughout evolution. Light dots represent amino acids that differ between the ancient and modern proteins. Image Credit: Dorothee Kern/Brandeis/HHMI.
Obsolete forms of two cell division proteins were recreated by biochemist Dorothee Kern and her team, who then observed the proteins in the lab as one promoted the function of the other. According to the study findings reported by the researchers in the February 21st issue of the journal Science, the observation represents the earliest known occurrence of two proteins interacting in such a manner.
The interaction is termed allosteric regulation and occurs between several modern proteins, but “how such a feature evolved is unexplored territory,” explains Kern.
Over the years, allosteric regulation has turned out to be a vital tool for cells. Kern explains that more refined controls are required to manipulate cells as they developed more complex networks of protein activity.
Allosteric regulation is arguably the biggest evolutionary step in the development of higher organisms.”
Dorothee Kern, Howard Hughes Medical Institute Investigator, Brandeis University
Kern intends to find how proteins and other molecules function together, both in real-time and at the atomic level. Identification of the behavior of proteins when they send cellular signals, initiate chemical reactions, or attach to other molecules could offer researchers a new path for drug discovery.
A protein named Aurora A was chosen by Kern for analysis since it is highly crucial in cell division—abnormalities can induce the growth of cancer—and since it is stimulated by a protein partner known as TPX2.
Aurora A helps the equal distribution of chromosomes between daughter cells during cell division. TPX2 attaches to an “allosteric site” of Aurora A—a spot other than the protein’s active site. Then, Aurora A is brought by TPX2 to the active site and the protein is switched into high gear. Kern intended to gain insights into the evolution of the partnership between the two proteins.
Organisms spread over the evolutionary tree synthesize certain versions of the Aurora A and TPX2 proteins. Versions of TPX2 and Aurora A from a range of species—such as humans, bacteria, and plants—were compared by Kern, study coauthor Adelajda Hadzipasic, and their collaborators at Brandeis.
Later, the researchers performed a bioinformatics analysis, traveling back in time to derive the age-old amino acid sequences of the oldest common ancestors of the two proteins. Then, those sequences were used to reconstruct ancient proteins.
“Evolution used to be speculative subject, but it is now experimental. That’s what’s really exciting about this technique,” stated Christopher Miller, an HHMI Investigator alum and recently retired biochemist who was Kern’s colleague at Brandeis
The interlinked histories of the proteins were mapped by Kern and her colleagues through experiments in the lab. Aurora A had to first self-activate nearly 1.5 billion years ago—when there was no need for TPX2. Almost a half billion years later, once TPX2 emerged, Aurora A’s productivity was increased by the protein by bringing it to the job site. “That created an evolutionary increase in fitness,” stated Kern.
According to Kern, the partnership lasted since Aurora A’s efficiency increased by partnering with TPX2, and the two proteins gained enough time to forge a more refined relationship.
As time passed, TPX2 assumed a new, additional role—that of Aurora A’s activator. At present, the latest TPX2/Aurora A pair can function 10 times faster compared to Aurora A alone.
Despite nearly a billion years of evolution between the two proteins, when Kern and her colleagues blended ancient Aurora A with modern TPX2, they discovered that both attached together strikingly well.
According to Kern, this was not predicted. Based on traditional perception, proteins that work together were assumed to evolve together—thus a change in one partner is balanced by a complementary change in the other. The team considered that modern TPX2 would likely have undergone many such changes to connect with such an ancient partner.
This mystery was solved by Kern. The team found that the sections of TPX2 and Aurora A that contact each other typically remained the same over a billion years, although other parts of the proteins deformed and shifted over time.
According to Neel Shah, a chemist at Columbia University, an in-depth study of such interactions could offer a new way to create new kinds of drugs. There is a huge gap between finding out the allosteric sites of proteins and actually designing drugs with the ability to take advantage of them, but “these findings could be a starting point,” he said.
Source:
Journal reference:
Hadzipasic, A., et al. (2020) Ancient origins of allosteric activation in a Ser-Thr kinase. Science. doi.org/10.1126/science.aay9959.