The mitochondrial genome (chondriome or mitogenome) and plastid genome (such as plastome, plastids including the chloroplast and other forms of plastid) denote the parts of endosymbiotic organelle inheritance in eukaryotes that have persisted in organelles without being lost or transferred to the nucleus.
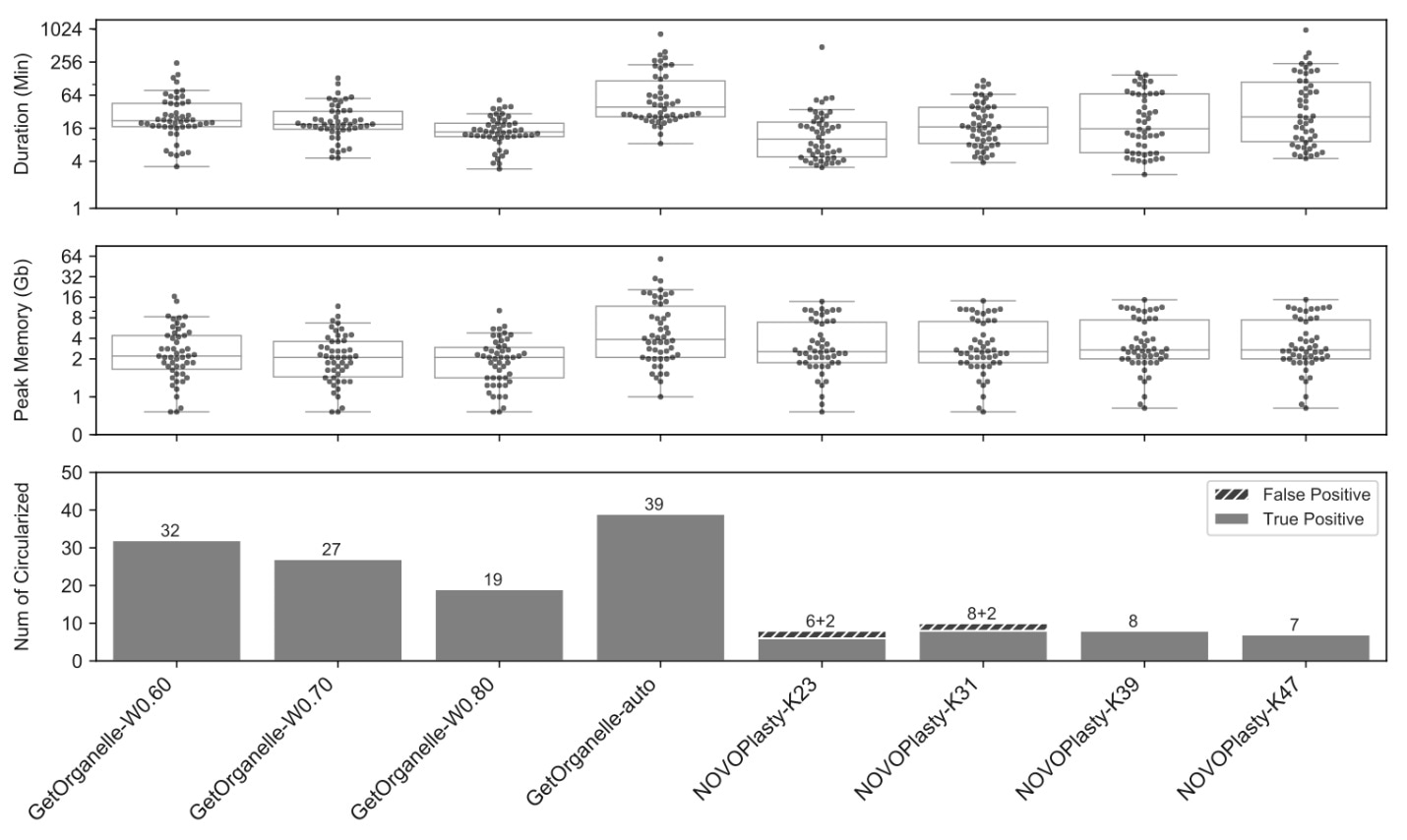
Comparisons of four sets of runs using GetOrganelle and four sets of runs using NOVOPlasty when assembling 50 public plant datasets. Image Credit: Kunming Institute of Botany.
The DNA sequences from the organelle genomes have been extensively employed in evolutionary and phylogenetic analyses, and also in DNA barcoding. Since high copy numbers of the organelle genome are present in a single cell, it is possible to get sufficient coverage from the low coverage whole genome sequencing (WGS) information to organize complete organelle genomes.
With the rapid advancements of high-throughput sequencing techniques, a considerable amount of WGS information was created in a cost-efficient manner, which, in turn, increases the need for precise and high-throughput assembly of organelle genomes.
While several pipelines and toolkits for assembling the organelle genomes have been created, the assembly qualities and efficiencies of these products are often below expectations.
A team of researchers, headed by Professor LI Dezhu and Professor YI Tingshuang from the Kunming Institute of Botany of the Chinese Academy of Sciences (KIB/CAS), has been engaged in comparative genomics, plastid phylogenomics, and DNA barcoding for several years.
Collectively, the taem has established a research system using the plastome information for evolutionary and phylogenetic studies, and achieved productive outcomes.
In addition, the teams were dedicated to design a novel tool for the analyzing plastome, such as a well-known plastome annotation toolkit called PGA.
Aiming at efficient and precise assembly of organelle genomes, an international collaborative team headed by Professor LI and Professor YI, with colleagues from KIB, the Xishuangbanna Tropical Botanical Garden of CAS, and Pennsylvania State University, has now designed GetOrganelle—a sophisticated toolkit for accurate de novo assembly of organelle genomes.
It is indeed a breakthrough that the GetOrganelle tool offers a pre-grouping approach for accelerating target-associated reads recruitment as well as contig multiplicity estimation algorithm for more improved repeat resolution. The novel algorithm of contig multiplicity estimation integrates both data of contig coverage and graph characteristics.
To assess the precision of assemblies using the GetOrganelle toolkit, in comparison to another well-known assembler called NOVOPlasty, the GetOrganelle research team examined 156 public datasets from fungi, animals, and plants.
For 50 public plant WGS datasets, the GetOrganelle toolkit created a high completeness rate of 78% with default settings, considerably better than the present most popular tool, NOVOPlasty, which created 16% with fine-tuning but cost somewhat less computational resources.
The GetOrganelle toolkit still considerably outpaced the NOVOPlasty tool in terms of the completeness rate even when consuming similar or less computational resources. Moreover, the NOVOPlasty tool created 20% to 25% wrong or false complete plastomes in K=23 and K=31 runs.
But in the same test, the consistency of GetOrganelle assemblies under varied parameters was also better than that of NOVOPlasty assemblies.
As per the read mapping assessment, GetOrganelle plastomes exceeded NOVOPlasty as well as published plastomes from the same reads in terms of accuracy.
Within the published plastomes, several mistakes were identified during this assessment. For 50 fungal datasets and 56 animal datasets, the GetOrganelle tool was usually better over NOVOPlasty tool when it comes to acquiring mitogenome genes and contigs.
Most significantly, in 2020, Freudenthal et al. presented a benchmark comparison of many chloroplast assembly toolkits/pipelines—including GetOrganelle, Fast-Plast, chloroExtractor, org.ASM, NOVOPlasty, and IOGA—and identified considerable variations among those assemblers.
In their tests, the GetOrganelle tool considerably outperformed all other assemblers in terms of precision and success rate, and was proposed as the default assembler.
Source:
Journal reference:
Jin, J-J., et al. (2020) GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology. doi.org/10.1186/s13059-020-02154-5.