A team of scientists headed by Professor Shouhong Guang from the University of Science and Technology of China (USTC) of the Chinese Academy of Sciences, associated with Professor Xuezhu Feng from the First Affiliated Hospital of USTC, disclosed the nucleolar RNA interference based on Caenorhabditis elegans (C. elegans) as a model organism.
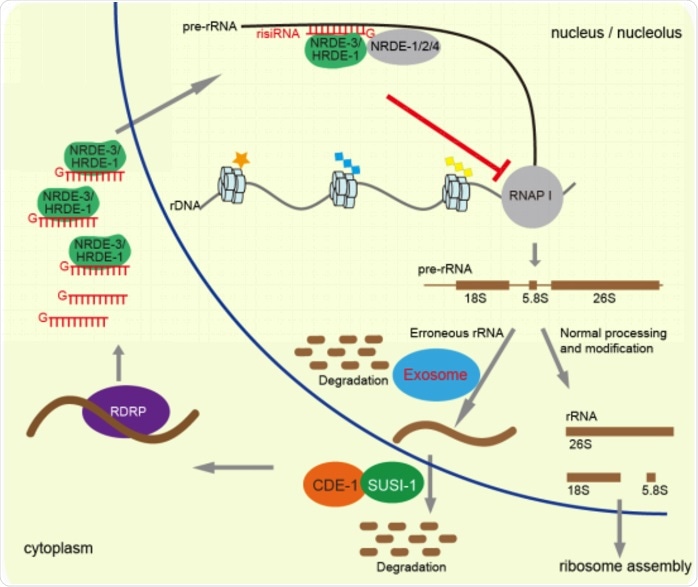
A working model of risiRNA biogenesis and function. Image Credit: Image by Shimiao Liao et al.
The research was published in the journal Nuclear Acids Research.
siRNA is a type of non-coding small RNA and is around 22 nucleotides long. siRNA is a great drug molecule and performs a vital role in the growth and development, reproductive genetics, and immune defense of both the plant and animal body.
Professor Guang’s team employed forward and reverse genetic screening and pinpointed a series of suppressor of siRNA (SUSI) factors that hinder endogenous siRNA production in C. elegans.
A defective SUSI factor results in the accumulation of misprocessed ribosomal RNA (rRNA) fragments in the cells, triggering the production of ribosomal siRNA (risiRNA). The risiRNAs complement rRNA sequences and control the level of rRNA by triggering the nuclear RNA interference pathway.
The scientists, in their research, cloned a SUSI-5 gene that inhibits the production of risiRNA through genetic screening and identified that the SUSI-5 gene encodes a conserved catalytic subunit DIS-3 of RNA exosomes.
The researchers employed CRISPR gene-editing technology and affirmed that the functional defect of any subunit of RNA exosome can lead to the aggregation of risiRNA. The risiRNA triggers two nuclear RNA interference key factors—NRDE-2 and NRDE-3—to infiltrate the nucleolus, attach ribosomal RNA precursors, and later hinder the transcriptional activity of RNA polymerase.
Furthermore, the scientists discovered that RNA exosomes can respond to the homeostasis of rRNA in the nucleus through the change of subcellular localization. RNA exosomes carry out a vital role in the processing and metabolism of various RNA and are majorly situated in the nucleolus.
The imbalance of intracellular rRNA homeostasis leads to the movement of RNA exosomes from the nucleolus to the nucleoplasm. The proper subcellular localization of RNA exosomes has a vital role in inhibiting the production of risiRNA.
The current research put forth a new rRNA monitoring model that enables individuals to comprehend the biological roles and processes of risiRNAs.
Source:
Journal reference:
Liao, S., et al. (2021) Antisense ribosomal siRNAs inhibit RNA polymerase I-directed transcription in C. elegans. Nucleic Acids Research. doi.org/10.1093/nar/gkab662.