Reviewed by Danielle Ellis, B.Sc.Jul 12 2023
The astounding variety of protein structures and their folds in nature has been further illuminated by a ground-breaking discovery. Researchers wanted to know how much of the huge range of potential protein structures nature had examined. The findings have broadened the understanding and revealed the depth of the protein cosmos by revealing an incredible variety of previously undiscovered protein folds.
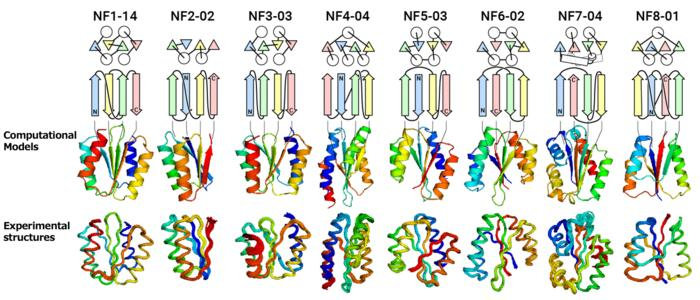 From the top, the protein name, topology diagram (circles represent α-helices, triangles, and arrows represent β-strands), computationally designed structures, and experimentally determined structures. The topology of NF8-01 forms a knot. Image Credit: Akira Yokoi
From the top, the protein name, topology diagram (circles represent α-helices, triangles, and arrows represent β-strands), computationally designed structures, and experimentally determined structures. The topology of NF8-01 forms a knot. Image Credit: Akira Yokoi
On July 3rd, 2023, this study was published in the journal Nature Structural and Molecular Biology.
The fundamental units of life, proteins, fold into particular three-dimensional shapes to carry out their biological activities. Proteins’ amino acid sequences determine their three-dimensional structures.
The structures of numerous proteins have been effectively revealed by experimental methods over time, but the occurrence of novel protein folds, which are determined by the configuration and connectivity of α-helices and β-strands, has decreased with time.
Image Credit: Christoph Burgstedt/Shutterstock.com
This begs the question, to what extent has nature not explored the protein fold space? Theoretical investigations have been carried out in an effort to answer this age-old query, yet there is a lack of experimental validation.
The research team started a project that combined theoretical prediction for novel protein folds with experimental confirmation of their de novo designs to answer this topic.
To theoretically forecast potential protein folds, the study team developed guidelines based on physical chemistry and protein structural data. Then, these guidelines were used to predict novel αβ-folds, which are made up of a four to eight stranded β-sheet and have not yet been seen in the Protein Data Bank (PDB).
This resulted in the discovery of 12,356 new folds in all. To test the foldability and accuracy of the predicted novel folds, the scientists then tried to computationally build proteins for them from scratch.
We attempted to computationally design proteins with all of the predicted folds that have a four-stranded β-sheet, including one forming a knot-like structure. When designing proteins, we did not expect all of them, especially knot forming ones, to fold into the structures as anticipated.”
Shintaro Minami, Researcher, Exploratory Research Center on Life and Living Systems
The findings of the testing were unexpected.
For all of the folds, the computationally designed protein structures closely matched the experimental structures.”
Naohiro Kobayashi, Senior Research Fellow. RIKEN
These findings imply that there are at least 10,000 undiscovered foldable αβ-folds, which is a noteworthy discovery given that just 400 αβ-folds have been seen in nature. This shows that there is still a great deal of undiscovered folds in the protein folding domain.
Several theories about the structure and evolution of proteins have been developed as a result of these findings. Proteins may not have been around long enough in biology for all potential folds to have been discovered, according to one theory.
Another theory holds that because all life on Earth descended from a single common ancestor, protein folds in nature are biased by nature.
Proteins may have evolved by repeatedly re-using specific folds while expressing different functions. If extraterrestrial life does exist, it might be utilizing a different set of protein folds.”
George Chikenji, Assistant Professor, Nagoya University
It is well recognized that proteins have a wide range of functions, which are derived from the variety of three-dimensional protein structures. This research has shown that there are at least 10,000 undiscovered foldable αβ-folds in nature.
Nobuyasu Koga, a professor at the Exploratory Research Center on Life and Living Systems (ExCELLS), National Institutes of Natural Sciences (NINS), concluded, “The design of proteins with these novel folds will lead to an even greater diversity of structures. This would pave the way for the de novo design of functional protein molecules, lead to breakthroughs in drug development, enzyme design, and other areas.”
Source:
Journal reference:
Minami, S., et al. (2023 Exploration of novel αβ-protein folds through de novo design. Nature Structural and Molecular Biology. doi.org/10.1038/s41594-023-01029-0