Reviewed by Danielle Ellis, B.Sc.Oct 16 2023
In the pursuit of sustainable agriculture, a key goal is to enhance nitrogen use efficiency (NUE) in crops. The extensive use of nitrogen (N) fertilizers since the 20th century has significantly boosted agricultural productivity. However, the excessive application of N fertilizers has given rise to severe environmental concerns and increased energy consumption.
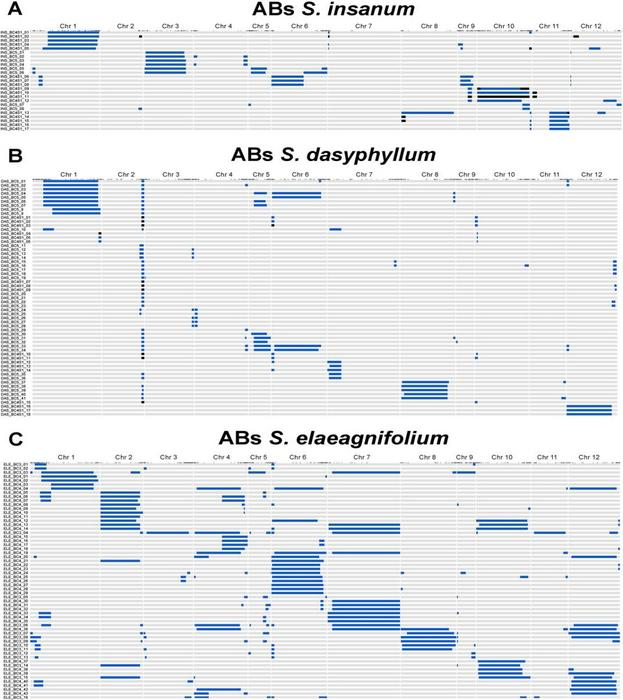 Graphical genotypes of ABs lines of S. insanum (A; n = 25), S. dasyphyllum (B; n = 59), and S. elaeagnifolium (C; n = 59) assessed for the present experiment. Image Credit: Horticulture Research.
Graphical genotypes of ABs lines of S. insanum (A; n = 25), S. dasyphyllum (B; n = 59), and S. elaeagnifolium (C; n = 59) assessed for the present experiment. Image Credit: Horticulture Research.
To tackle this challenge, crop wild relatives (CWR) offer valuable genetic resources that can be harnessed in breeding programs. Among the wild relatives of eggplant (Solanum melongena L.), there are three categories: primary (GP1), secondary (GP2), and tertiary (GP3) gene pools, which represent underexplored genetic resources.
Nevertheless, the direct utilization of crop wild relatives (CWRs) in breeding is challenging due to inherent genetic barriers. This highlights the critical need for the development and investigation of advanced backcrosses (ABs) to efficiently integrate these advantageous traits.
In July 2023, the study was published in Horticulture Research.

Image Credit: nnattalli/Shutterstock.com
In the current study, 22 morpho-agronomic, physiological, and NUE traits were assessed under low nitrogen (LN) fertilization conditions in CWRs of eggplant (S. insanum, S. dasyphyllum, and S. elaeagnifolium) and their advanced backcrosses (ABs; BC3 to BC5 generations).
Genome coverage of the donor wild relatives differed and the highest coverage was seen in S. elaeagnifolium at 99.2%. Significant representation was noticed on chromosomes 1 (86.8%) and 3 (80.9%) in S. insanum, while for S. dasyphyllum, the emphasis was on chromosomes 1 (84.8%) and 5 (86.3%).
While characterizing S. melongena recurrent parents (MEL5, MEL1, and MEL3), noticeable disparities were observed between nitrogen treatments. For example, a 3.7-fold and 5.0-fold difference in yield and fruit number (F-Number), respectively, was noticed across treatments for MEL5.
Moreover, fruit metrics, like fruit pedicel length in MEL5, showed variations under differing nitrogen conditions. Principal components analysis (PCA) disclosed trait groupings among the AB sets, with 48.8% total variation accounted for in the S. insanum and its recurrent parent S. melongena MEL5.
Pearson linear correlations revealed noteworthy associations among traits across the advanced backcross (AB) sets. A total of 16 potential quantitative trait loci (QTLs) were detected within the AB sets, suggesting genetic regulation of specific traits, and potential candidate genes were identified from the eggplant reference genome assembly.
Among the 16 potential quantitative trait loci (QTLs) that were identified, five were found to be clustered at the same position on chromosome 9 of S. insanum. The '67/3' eggplant reference genome provided additional insights into potential candidate genes, such as the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER located on chromosome 9.
In summary, this study underscores the significant potential of eggplant wild relatives for genetic enhancement under low nitrogen conditions, thereby advancing the cause of sustainable agriculture. The identified quantitative trait loci (QTLs) and their relationships establish a foundation for innovative eggplant breeding initiatives aimed at enhancing yield, quality, and nitrogen use efficiency under low nitrogen conditions.
Source:
Journal reference:
Villanueva, G., et al. (2023). Evaluation of three sets of advanced backcrosses of eggplant with wild relatives from different gene pools under low N fertilization conditions. Horticulture Research. doi.org/10.1093/hr/uhad141.