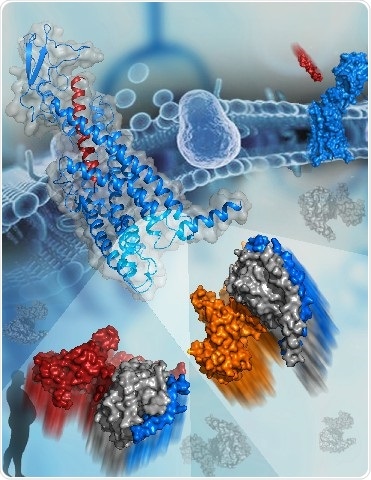
The active structure of GCGR bound to glucagon (blue and red cartoon, top left corner). The two G proteins that bind to GCGR are shown as surfaces at the bottom. The subunits of Gs are colored orange, gray, and blue. The subunits of Gi are colored red, gray, and blue. Image Credit: Dr. HAN Shuo.
The biological action of the receptor depends largely on the selective coupling of a GPCR and particular G proteins.
Insights into the mechanisms of GPCR signal transduction have been limited since the molecular details characterizing how an individual GPCR identifies different G protein subtypes remain elusive.
Two cryo-electron microscopy (cryo-EM) structures of the human glucagon receptor (GCGR) together with its relative agonist glucagon and different classes of G proteins, Gs or Gi, were determined by a research team headed by WU Beili and ZHAO Qiang at the Shanghai Institute of Materia Medica (SIMM) of the Chinese Academy of Sciences (CAS), a team led by SUN Fei at the Institute of Biophysics of CAS, and a team guided by Denise Wootten from Monash University.
The study was published on March 20th, 2020, in the Science journal.
A comprehensive molecular map of patterns of interaction between a GPCR and different G protein subtypes is provided by these structures, for the first time, surprisingly revealing several molecular features that control G protein specificity. This significantly improves the insights into GPCR signaling mechanisms.
GCGR belongs to the class B GPCR family and is vital for glucose homeostasis by initiating the release of glucose from the liver. This makes it a prospective drug target for obesity and type 2 diabetes.
GCGR canonically applies its physiological action via Gs signaling, yet it can bind to other G proteins such as Gi and Gq, thus enabling diverse cellular responses.
The crystal structures of the full-length GCGR coupled to a negative allosteric modulator or a partial peptide agonist was identified by researchers at SIMM in 2017 and 2018. That study offered an understanding of signal recognition and modulation of class B GPCRs.
Now, the researchers have achieved further progress by resolving the complex structures of GCGR attached to two transducer proteins with contradictory biological activities. This research offers a valuable understanding of pleiotropic GPCR-G protein coupling and G protein specificity.
Most importantly, it showed that GCGR’s sixth transmembrane helix (helix VI) goes through a similar outward shift in the two G protein-bound GCGR structures, thereby creating a common binding cavity to house Gs and Gi.
This is in contrast to the theory based on the previously identified GPCR-G protein complex structures, which hypothesized that the difference in positions of helix VI is a crucial determinant in the coupling specificity of Gs and Gi.
The common G protein coupling pocket visualized in the complex structures of the GCGR-G protein is consistent with the signaling pleiotropy of GCGR and enables ultimate efficiency in the activation of different pathways.
This common pocket is used by GCGR to bind to both G proteins, but GCGR performs this action in various interaction patterns accounting for G protein specificity. The quantified interaction interface between Gs and GCGR is considerably larger than it is for Gi, which leads to the increased binding affinity of Gs to the receptor. This provides a structural foundation for the preferential binding of GCGR to Gs.
The researchers took the structures of GCGR-Gs and GCGR-Gi complexes as a basis and carried out wide-scale functional studies with the help of techniques such as G protein activation, mutagenesis, and cell signaling to analyze the roles of main residues in the receptor-G protein coupling interface during Gs and Gi activation.
The study outcomes reveal that conformational variations of residue side chains and intracellular loops in the receptor are enough to guide G protein selectivity. The interactions brought about by the helix VII/VIII junction and the second intracellular loop (ICL2) of the receptor have a critical role in Gs coupling.
By contrast, ICL1 and ICL3—the other two intracellular loops—and the receptor hydrophobic intracellular binding cavity are highly crucial for Gi recognition.
These results offer in-depth knowledge about pleiotropic coupling, GPCR activation, and G protein specificity. They also offer new opportunities for drug discovery by developing biased ligands for the selective inhibition of one particular signaling pathway, which leads to reduced side effects.
Source:
Journal reference:
Qiao, A., et al. (2020) Structural basis of Gs and Gi recognition by the human glucagon receptor. Science. doi.org/10.1126/science.aaz5346.