However, it is not clear which interactions safeguard health and which ones can lead to disease when they go amiss.
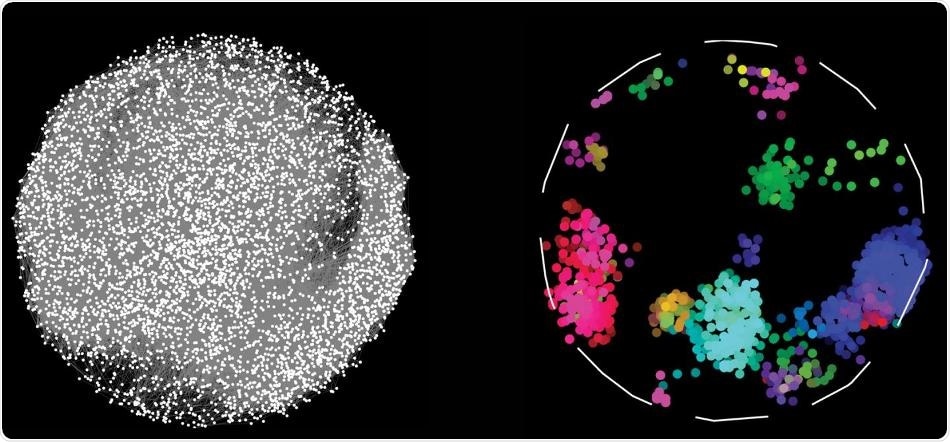
By charting pairwise interactions between 17,500 human proteins, scientists have created a map, on the left, depicting which proteins work together to sustain cellular function. Proteins with similar interactions profiles fall into discrete color-coded clusters representing different bioprocesses in the cell. Image Credit: Katja Luck et al.
The human genome project has offered a “parts list” for the cell. However, only an in-depth understanding of how these parts work together or interact can offer insights into the inner workings of the cell and what goes wrong in the case of a disease.
To resolve these questions, researchers needed a reference map of interactions, or an interactome, between gene-encoded proteins, which constitute cells and perform a majority of the work in them.
Since the mid-1990s, our collaborative team has pushed the idea that interactome maps can illuminate fundamental aspects of life. Our paper describes the first human interactome reference map, constituting a ‘scaffold’ of information to better understand how faulty genes cause diseases such as cancer, but also how viruses such as the coronavirus that causes COVID-19 interact with their host human proteins.”
Marc Vidal, Team leader and Director, Center for Cancer Systems Biology (CCSB), Dana-Farber Cancer Institute
The human protein map has been in the making for nearly 10 years. It is now available as a result of a collaborative effort, which involved more than 80 researchers in the United States, Canada, Spain, Belgium, France, and Israel, jointly headed by Vidal, David E Hill, and Michael A Calderwood, at Dana-Farber Cancer Institute, as well as Frederick P Roth, at the University of Toronto’s Donnelly Centre for Cellular and Biomolecular Research.
A study reported in Nature describes that the Human Reference Interactome (HuRI) map is the largest of its kind and charts 52,569 interactions between 8,275 human proteins.
Humans have nearly 20,000 protein-coding genes; however, to date, researchers know far less about a majority of the proteins they encode. Luckily, it is possible to gather this information from interaction data with the help of the “guilt by association” principle.
As per this principle, it is likely that two proteins with similar interacting partners are involved in analogous biological processes.
We can use our human interactome map to predict protein function. People can look up their favourite protein and get clues about its function from the proteins it interacts with.”
Frederick P Roth, Donnelly Centre for Cellular and Biomolecular Research, University of Toronto
Roth is also a Senior Scientist at the Sinai Health System’s Lunenfeld-Tanenbaum Research Institute. The data are already offering crucial understanding about, for instance, new cellular roles for human proteins and what goes wrong at the molecular level to spur on the disease.
In this regard, HuRI has already divulged new functions for proteins that play a role in programmed cell death, discharge of cellular cargo, and other processes.
The researchers combined tissue-specific gene expression with protein interaction data, thus determining the protein networks that play a role in the formation and maintenance of different tissues, unveiling new therapeutic targets for different genetic diseases, such as cancer, and prospectively even for infectious diseases.
In addition, the researchers made use of HuRI as a reference to observe how disease-causing protein variants produce network rewiring to show the molecular mechanisms that cause those specific disorders.
Genome sequencing can identify the variants carried by an individual that make them susceptible to disease, but it doesn’t reveal how the disease is caused. Changes in the interactions of a protein is one possible mechanism of disease, and this map provides a starting point to study the impact of disease associated variants on protein-protein interactions.”
Mike Calderwood, PhD, Scientific Director, Center for Cancer Systems Biology (CCSB), Dana-Farber Cancer Institute
Earlier, the Toronto and Boston researchers performed two smaller studies that mapped approximately 14,000 protein interactions in total. Currently, HuRI has investigated proteins encoded by almost all human protein-coding genes and widened the map four-fold.
The teams created HuRI by co-expressing in pairs nearly all human proteins in yeast cells. Upon interacting, or binding with one another, the two proteins form a molecular switch that promotes the growth of yeast cells, which indicates that there has been an interaction.
Of 17,500 proteins, the researchers tested all probable pairwise combinations for their potential to interact with one other in three individual versions of a yeast-based assay. Each one was performed in triplicate, amounting to an overwhelming three billion separate tests.
The results produced nearly 53,000 high-confidence binary interactions between more than 8,000 proteins, which were validated using other techniques. Most of the interactions had not been detected earlier.
This map is the largest of its kind created until now, but it is still incomplete and represents about 2%–11% of all human protein interactions. According to Roth, one reason for many interactions being missed could be the lack of some human-specific molecular factors in yeast cells, which are required for proper protein function.
In spite of these drawbacks, HuRI has increased over three times the number of known interactions between human proteins and will be a crucial resource for the research community.
The data web portal, which was developed by Miles Mee, Mohamed Helmy, and Gary Bader (also in the Donnelly Centre), has already been visited by 15,000 people since HuRI was made accessible on bioRxiv, an open-source online publisher, in April 2019.
“We already had lots of people download the whole dataset and so I imagine we’ll see the iteration of our previous paper, which has already been cited over 800 times and is less than a third of the size of HuRI,” added Roth.
Source:
Journal reference:
Luck, K., et al. (2020) A reference map of the human binary protein interactome. Nature. doi.org/10.1038/s41586-020-2188-x.