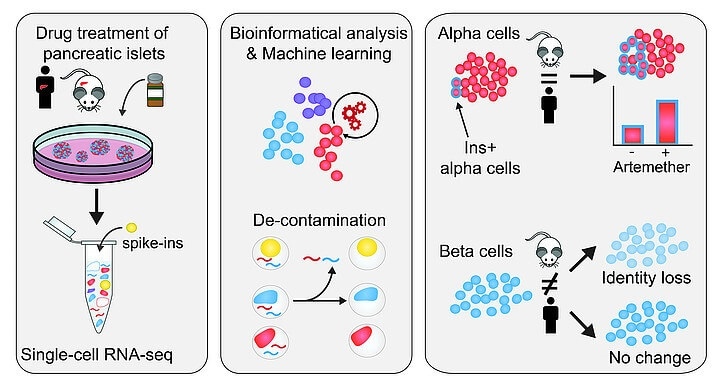
The workflow of identification and removal of contaminating reads in single-cell sequencing and the islet cell type-specific drug responses of the decontaminated profiles Image Credit: © Brenda Marquina-Sánchez and Nikolaus Fortelny/CeMM.
Such results will assist the development of targeted drug therapies for treating Type 1 diabetes in the days to come.
The pancreas can be described as an abdominal organ that releases digestive enzymes and also hormones that control blood sugar levels. This hormone-producing organ is located in the islets of Langerhans, which is made up of clusters of different types of endocrine cells.
Among those clusters are beta cells, which release the hormone insulin required to reduce the blood glucose (a kind of sugar) levels in humans, and also alpha cells, which produce the hormone glucagon that is in charge of increasing the blood glucose levels.
Type 1 diabetes is a chronic disorder where the immune system of the body wrongly attacks and damages the insulin-producing beta cells of the pancreas.
The objective of regenerative medicine is to replenish the mass of beta cells and thus support and eventually replace the existing insulin replacement treatments.
Modifications to the composition of islet cells, such as inadequate beta-cell dedifferentiation and beta-cell function, also account for type II diabetes. Hence, a better interpretation of the crosstalk and identity of the different types of islet cells leads to a more improved characterization of both forms of diabetes and may result in the development of unique therapeutic ideas.
Single-cell transcriptomics is known to be a robust method to define cellular identity. Earlier, scientists from Christoph Bock’s and Stefan Kubicek’s teams at CeMM had published the initial single-cell transcriptomes from primary human pancreatic islet cells.
Since then, technological advancements have allowed its application for creating global human and mouse single-cell transcriptome atlases.
But in spite of these developments, single-cell methods continue to remain technologically difficult, considering that the presence of the tiny amount of RNAs is fully used up in the experiment. Hence, the purity and quality of the ensuing single-cell transcriptomes should be ensured.
At the two contributing laboratories, CeMM scientists detected unexpectedly high expression of the hormone in non-endocrine cell types, in their individual dataset and also in other published single-cell studies.
The scientists endeavored to explain whether this would be the outcome of contamination caused by RNA molecules, for instance, from dying cells, and how it could be eliminated to obtain a more consistent dataset. Such contamination appears to occur in single-cell RNA-seq data from a majority of the tissues, yet it was highly visible in pancreatic islets.
Islet endocrine cells are solely devoted to the production of single hormones, and glucagon in alpha cells and insulin in beta cells are highly expressed than the characteristic “housekeeping” genes.
Therefore, redistribution of these transcripts to other types of cells was extremely pronounced. On the basis of this observation, the researchers’ aim was to create, verify, and apply a technique to find out, through an experiment, and computationally eliminate such contamination.
In this study, CeMM scientists employed spiked-in cells from different types of cells, mouse as well as human samples, and added these to their samples of pancreatic islet cells. Most significantly, the transcriptomes of these spike-in cells were completely defined.
This enabled the team to manage the level of RNA contamination in single-cell RNA-seq both internally and precisely, considering that the human transcripts identified in the mouse spike-in cells make up the contaminating RNA.
In this manner, the researchers discovered that the samples had a contamination level of around 20%, and thus effectively defined the contamination in individual samples. Then, they developed a unique bioinformatics method to eliminate the contaminating reads from single-cell transcriptomes computationally.
Having achieved a “decontaminated” transcriptome from which the false signal was removed, the team went on to define how the cellular identity in different types of cells reacted to the treatment with three various drugs.
The researchers discovered that a small molecule inhibitor of the transcription factor FOXO1 promotes dedifferentiation of alpha cells and beta cells. They also examined the artemether, which had been identified to decrease the function of alpha cells and can potentially induce insulin production in in-vitro and in-vivo studies. (The effects of the artemether drug were both cell-type-specific and species-specific.)
In alpha cells, a few cells increase the expression of insulin and gain aspects of the identity of beta cells in both mouse and human samples.
Significantly, the scientists observed that human beta cells did not show any major change in insulin expression, while in mouse islets, beta cells reduced their insulin expression and also the overall identity of the beta cells.
Source:
Journal reference:
Marina-Sanchez, B., et al. (2020) Single-cell RNA-seq with spike-in cells enables accurate quantification of cell-specific drug effects in pancreatic islets. Genome Biology. doi.org/10.1186/s13059-020-02006-2.