The neurotransmitter dopamine plays a vital role in everything, starting from motor control to higher cognitive functions, arousal, motivation, sexual gratification, and reinforcement.
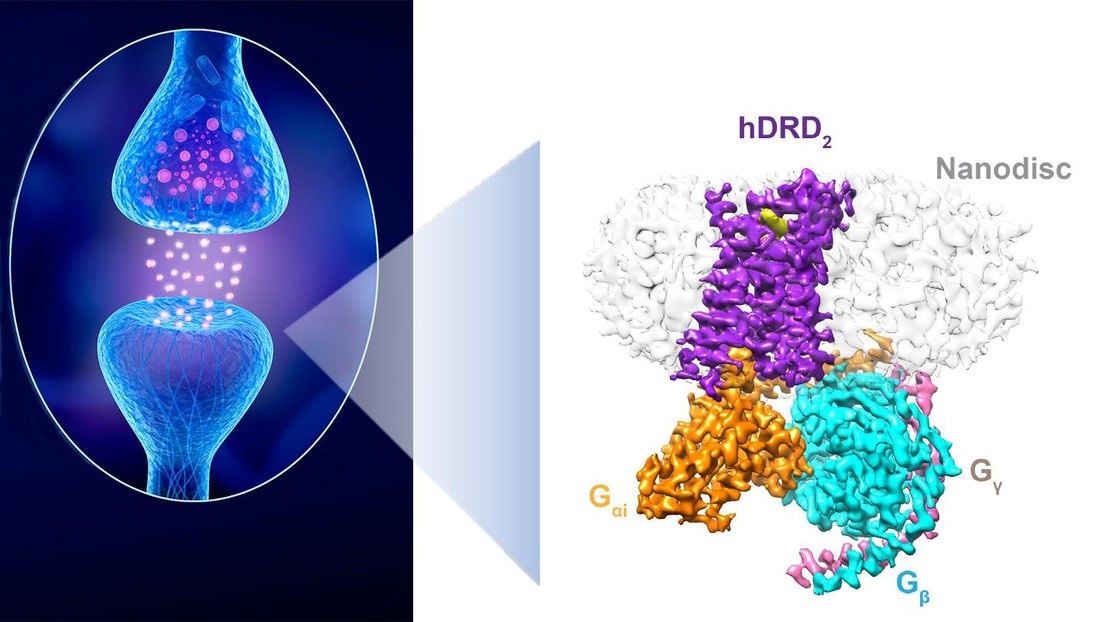
A dopaminergic synapse between a neuron releasing dopamine and a neuron sensing the neurotransmitter. Right: The structure of a D2 receptor in “action.” Image Credit: iStock and P. Barth (EPFL).
Dopamine acts on receptors that have a longstanding target for treating several disorders including Parkinson’s disease, which results from the degeneration of dopamine-using neurons that regulate movements.
The major drawback is that for a span of 20 years, none has been able to “see” the structure of a dopamine receptor when it is triggered by dopamine—at least not in sufficient resolution to create opportunities for designing drugs that can effectively target the receptors.
In a prominent collaborative study published in the Nature journal, researchers from the laboratory of Patrick Barth at Ecole Polytechnique Fédérale de Lausanne (EPFL) and collaborators at UCSD and UTSW have recently created a high-resolution structure of a stimulated form of a dopamine receptor in a native lipid membrane setting.
The native receptor is so misbehaved and its active form so transient that attempts at observing the receptor structure ‘in action’ have failed so far.”
Patrick Barth, EPFL
The way the researchers solved the issue by integrating advanced computational allosteric and de novo protein design strategies developed by Barth’s team allowed them to engineer a highly stable but stimulated dopamine receptor, the structure of which could then be studied and resolved.
The EPFL researchers produced a receptor with artificial building blocks, like de novo binding sites and activating switches, which could replace the structurally disordered, unstable, and inactivating areas of the native receptor.
This hybrid functional/de novo computational protein design approach is powerful, as it enabled us to create a receptor with considerably enhanced activity and stability while recapitulating key native functionalities such as dopamine-mediated intracellular signaling and binding.”
Patrick Barth, EPFL
The researchers achieved success by using cryo-electron microscopy and high-end lipid reconstitution techniques, overcoming barriers in the earlier studies that tried to determine the structure of the receptor using X-ray crystallography and by retaining the receptor within detergents.
The disadvantage is that the detergents poorly mimic the lipid membranes of the cell, where receptors like the dopamine are naturally situated. Detergents are also known to deform and inactivate receptors, and this fact does not help when attempting to observe what they look like in action.
This represents the first atomic-level membrane-receptor structure determined in a native lipid bilayer. The breakthrough will enable improved drug discovery efforts against, for example, Parkinson disease.”
Patrick Barth, EPFL
“But it also sets the stage for broadly applying functional and de novo protein design approaches to accelerate the structure determination of challenging protein targets and create proteins with novel functions for a broad range of therapeutic and biotechnological applications,” Barth concluded.
Source:
Journal reference:
Yin, J., et al. (2020) Structure of a D2 dopamine receptor-G protein complex in a lipid membrane. Nature. doi.org/10.1038/s41586-020-2379-5.