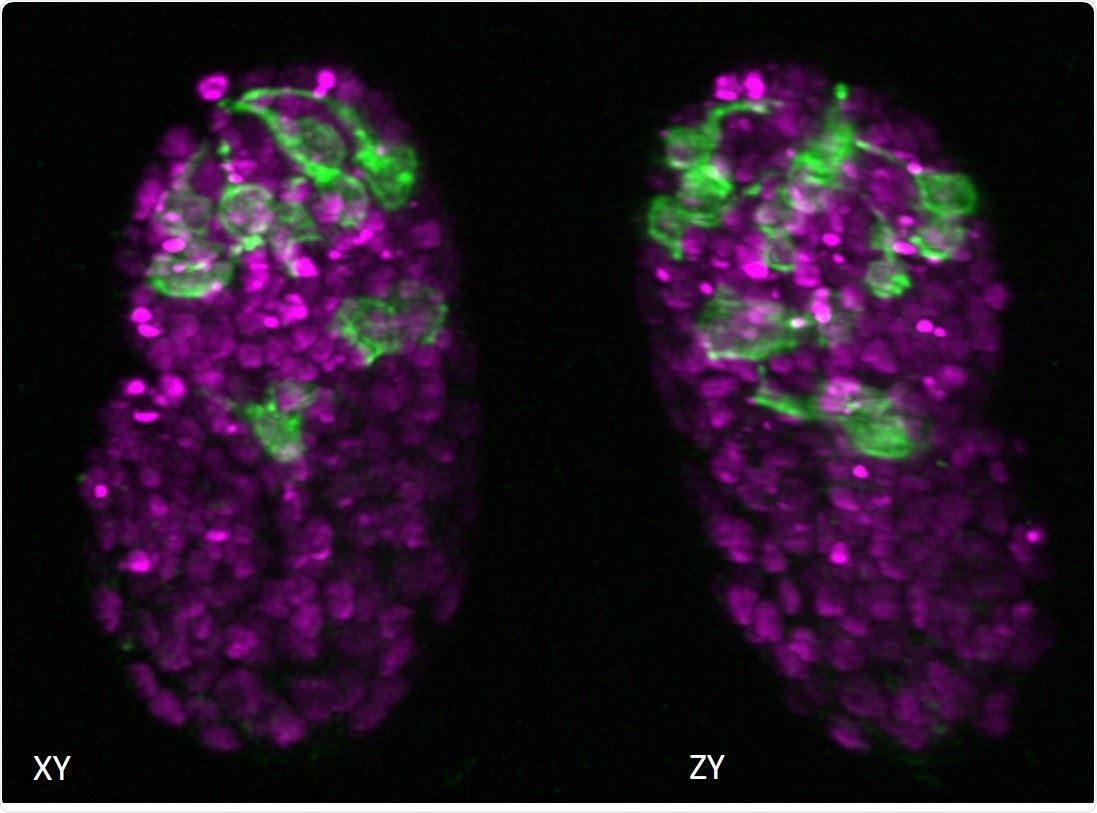
Two views of a C. elegans embryo. The green fluorescence shows the neurons and the magenta areas are the nuclei of the cells. Image Credit: National Institute of Biomedical Imaging and Bioengineering.
Microscopes like these are generally used to observe the neural development of worm embryos, capture brain cells and blood cells that move through fish, and locate individual organelles inside the whole organs.
With microscopes becoming increasingly better and producing higher resolution images more rapidly, scientists have to deal with more amounts of data and have less processing time. Although the videos themselves can be recorded in minutes, the generated images are likely to be terabytes in size and may need weeks or, in certain cases, months of usable processing time.
One of the reasons for this time-consuming process is that the videos generally record small objects that are distorted by the microscope. Such blurring effects can be minimized by a process known as deconvolution, but it requires plenty of time and computing power.
Another problem is that a majority of the microscopes being used today take numerous views of the same cell or organism. Such images should be placed correctly and subsequently merged to create three-dimensional (3D) images and videos.
Extensive computer processing time is required to produce high-resolution images from the raw data. Therefore, while the microscopes have been advanced to offer high-resolution and increasingly complex images, computing power has restricted the types of techniques that are practical for scientists to use since they already know that most of the data collected by them will remain unused.
Shroff’s laboratory and his colleagues initially tried to alter the deconvolution algorithm employed by several scientists, so that it can run faster. This method, first suggested for other fields of medical imaging, like computed tomography (CT), was effectively modified for use with fluorescence microscopy, for the first time.
Fluorescence microscopy employs dyes to enhance contrast in the specimen, enabling scientists to target particular parts of a sample and observe how various elements communicate with one another.
Secondly, the scientists decreased the time required to place and stitch numerous views of a sample together. A major part of this development depended on a process known as parallelization. It is a method that is occasionally employed in supercomputing where rather than processing every individual function consecutively, the task is split into smaller ones that can be examined simultaneously.
It is similar to asking scores of persons to solve a single math problem individually, instead of asking a single person to solve an unlimited number of problems.
The scientists ultimately demonstrated that they could further decrease the time required to process the data through a neural network—a form of artificial intelligence (AI). Today, AI is increasingly used to assist diagnoses and image processing. In this example, Shroff and his research team trained the neural network to create higher and cleaner resolution images much more quickly than otherwise would be possible.
Acquiring modern imaging data is a bit like drinking from a firehose. These methods help us obtain valuable biological information faster, which is essential, given the massive amount of data that can be produced by these microscopes.”
Hari Shroff, PhD, Chief of Laboratory on High Resolution Optical Imaging, National Institute of Biomedical Imaging and Bioengineering
Such advancements extend the use of prevailing technology, such as facilitating the imaging of dense samples that create large amounts of image data when analyzed with fluorescence microscopes.
The advancements are equally important for using an increasing number of “computational microscopes” where the post-processing of incomprehensible raw data is a crucial step in creating the ultimate high-resolution image.
Shroff and his colleagues believe that they will assist scientists with methods they would not have assumed to try since it would be too labor-intensive to produce meaningful images.
Researchers from Harvard University, Sloan Kettering Institute, Yale University, University of Maryland, and the University of Connecticut also contributed to this study.
Source:
Journal reference:
Guo, M., et al. (2020) Rapid image deconvolution and multiview fusion for optical microscopy. Nature Biotechnology. doi.org/10.1038/s41587-020-0560-x.