Scientists from Harvard University, the Centro Nacional de Análisis Genómico (CNAG), and the Centre for Genomic Regulation (CRG) have demonstrated a pioneering technology that helps observe hundreds or perhaps thousands of genomes simultaneously under the microscope.
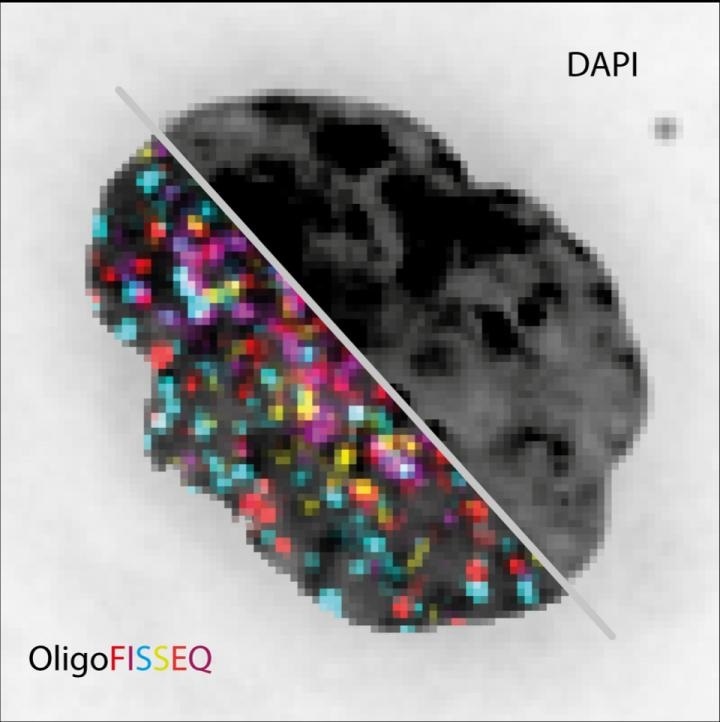
The image shows the same cell nucleus visualized with conventional fluorescence image (DAPI) compared to OligoFISSEQ for 129 different loci. Image Credit: Marc Marti-Renom.
The technology images genomes more quickly, more economically and improves the range of visibility when compared to presently available techniques. The method has been explained in the Nature Methods journal.
Every human cell contains 2 m of genome that is narrowed down to 10 µm inside the nucleus of the cell. Through this blueprint of life, genes are able to make physical contact with other kinds of genes that could be situated quite remotely along the chromosome.
A three-dimensional (3D) organization like this is important for cellular function; however, its constant dynamism and complexity make it extremely hard to visualize. To date, it has been impossible to image more than a handful of genes simultaneously, restricting the ability of scientists to define the function of genomes.
Fluorescence in situ hybridization (FISH) is one of the most standard methods used for analyzing the genome. This method employs fluorescent probes to label the absence or presence of particular sequences of DNA on chromosomes.
Thanks to the FISH technique, researchers have made breakthrough discoveries, for example, the division of cells. This technique is still being used today for species identification or medical applications, among other types of applications.
FISH is an aging technology that was initially developed in the 1980s, and hence, it can only observe a few genes simultaneously. Since that time, novel techniques have been developed that are capable of imaging genomes at a high resolution; however, these methods map an extremely reduced number of chromosomes or regions at a time.
The study’s authors have recently described a new technology called OligoFISSEQ in the Nature Methods journal. This technology uses new computational techniques that solve these restrictions. The researchers used this technology to produce 3D maps of 66 genomic targets over six chromosomes in an unlimited number of cells, displaying the ability of OligoFISSEQ to observe the complete genome at a molecular resolution that is beyond reach, to date.
Up to this moment, seeing a large number of different genes at the same time under the microscope was impossible. We combined existing sequencing technologies in a smart way so that we can see hundreds of genes by sequencing their targets under the microscope.”
Marc A. Marti-Renom, Study Co-Lead Author and ICREA Professor, Centro Nacional de Análisis Genómico-Centre for Genomic Regulation
Marti-Renom continued, “Before OligoFISSEQ, reaching this number of genes at the same time was slow and expensive. It is like upgrading from a dial-up phoneline to fiber-optic internet and paying 40 times less for it.”
In addition, the scientists tested the OligoFISSEQ technology by plotting all the 46 regions along the length of the human X chromosome. This technology has a higher resolution to unravel novel patterns in the way the genome arranges itself, for example, the length of the chromosome’s arm is not associated with its angle. The scientists theorized that this may be indicative of cell state, cell type, age, or cellular health.
The resolution offered by OligoFISSEQ has huge potential to shed new light on patterns we could not see before. Because of the coverage it provides to study the genome, it is well suited for spotting what may seem like a minor, seemingly inconsequential change in one part of the nucleus that may have a ‘butterfly effect’ elsewhere. What may have previously have been thought of as random may turn out to be anything but. That is the power of this tool.”
Marc A. Marti-Renom, Study Co-Lead Author and ICREA Professor, Centro Nacional de Análisis Genómico-Centre for Genomic Regulation
Source:
Journal reference:
Nguyen, H. Q., et al. (2020) 3D mapping and accelerated super-resolution imaging of the human genome using in situ sequencing. Nature Methods. doi.org/10.1038/s41592-020-0890-0.