An international group of researchers from Japan, the United States, and the United Kingdom studied the role of the histone demethylase KDM5A in multiple myeloma, one of the three main hematological cancers, and discovered the mechanism by which it stimulates myeloma cell proliferation.
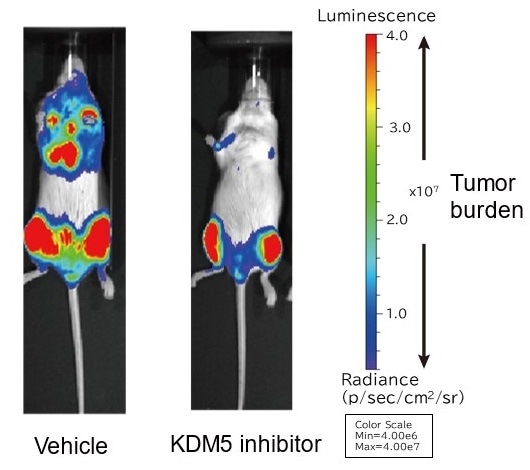
The ability of myeloma cells to proliferate was tested in a myeloma mouse model. Results show that the KDM5 inhibitor impaired myeloma cell proliferation. Image Credit: Associate Professor Hiroto Ohguchi.
In addition, they created a novel KDM5 inhibitor and demonstrated that it prevents cancer cell development in a myeloma mouse model. The researchers anticipate that new KDM5A-targeting therapies will be created in the future.
With the advent of new therapeutic agents each year, the prognosis for multiple myeloma improves, but there is also no cure. More research into the pathogenesis of this cancer, as well as the development of therapeutic agents, is expected.
The molecular pathogenesis of cancers, like multiple myeloma, is intricately linked to both epigenetic and genetic modifications. The epigenetic regulator KDM5 family proteins are abundant in myeloma cells, but their function remains unknown.
Researchers from Kumamoto University and the Dana-Farber Cancer Institute used genetic manipulation to inhibit the expression of KDM5 in human myeloma cell lines in order to better understand the function of KDM5 family proteins in myeloma cells.
They discovered that KDM5A, in particular, had a strong impact on cell proliferation, prompting them to investigate its molecular mechanisms. They also produced a novel KDM5 inhibitor and tested its effectiveness in both myeloma patient cells and mouse models.
Myeloma cell development was hindered by genetic repression of KDM5A expression or pharmacological inhibition of KDM5. Furthermore, the researchers showed that their KDM5 inhibitor inhibits myeloma cell development even in vivo by using myeloma mouse models in which a human myeloma cell line was transplanted into immunodeficient mice.
KDM5A collaborates with MYC, an essential transcription factor in myeloma growth and development, to stimulate the expression of MYC target genes, according to functional studies.
Since a high level of histone methylation (H3K4me3) was initially observed near the transcription start sites of MYC target genes, repressing KDM5A enhanced this modification level even further.
This suggests that excess H3K4me3 serves as a barrier to transcription, inhibiting transcription, in contrast to prior hypotheses that H3K4me3 facilitated transcription. Further investigation revealed that KDM5A assists transcription-associated complexes in moving from transcription initiation to transcription elongation by transiently releasing H3K4me3.
This research suggested a new model of epigenetic control in which KDM5A controls histone methylation at the transcription start site to optimal levels during the required phase, facilitating MYC target gene transcription and contributing to myeloma cell proliferation. It was also discovered that KDM5 inhibitors suppress myeloma cell growth.
Our research has elucidated part of the mechanism of myeloma cell proliferation mediated by histone modification regulation and has shown the potential for therapies targeting KDM5A. It is also becoming clear that the KDM5 family is involved in the growth of other carcinomas. The problem with the KDM5 inhibitors so far is that they have weak cell membrane permeability and are not effective in cells or in vivo.”
Hiroto Ohguchi, Study Lead and Associate Professor, Kumamoto University
“However, if a therapeutic drug is developed based on the KDM5 inhibitor developed in this study, it is expected to develop into a new therapeutic strategy for not only multiple myeloma but also various other types of cancer by combining it with conventional treatment methods,” concluded Ohguchi.
Source:
Journal reference:
Ohguchi, H, et al. (2021) Lysine Demethylase 5A Is Required for MYC-Driven Transcription in Multiple Myeloma. Blood Cancer Discovery. doi.org/10.1158/2643-3230.BCD-20-0108.