Proteases, one of the most inventive products, are utilized to enhance the efficacy of feed additives. Owing to their hydrolytic properties, they improve the absorption of amino acids, which can lessen the content of protein and the expense of feeds.
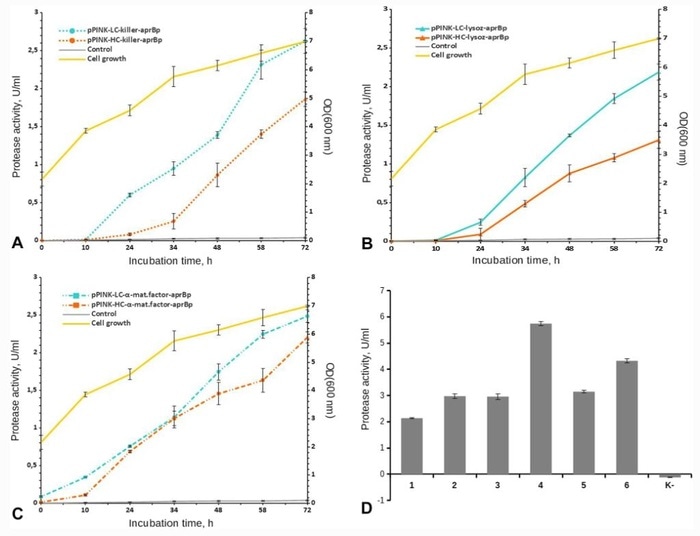
Dependence of proteolytic activity of subtilisin-like protease on the growth dynamics of P. pastoris recombinant yeast. a pPINK-HC-lysoz-aprBp and pPINK-LC-lysoz-aprBp; (b) pPINK-HC-killer-aprBp and pPINK-LC-killer-aprBp; (c) pPINK-HC-α-mat.factor-aprBp and pPINK-LC-α-mat.factor-aprBp; (d) determination of the specific activity of subtilisin-like protease by the hydrolysis of Z-Ala-Ala-Leu-pNa. 1, pPINK-HC-lysoz-aprBp; 2, pPINK-LC-lysoz-aprBp; 3, pPINK-HC-killer-aprBp; 4, pPINK-LC-killer-aprBp; 5, pPINK-HC-α-mat.factor-aprBp; 6, pPINK-LC-α-mat.factor-aprBp; K, culture fluid of recombinant yeast, before the addition of methanol. Image Credit: Kazan Federal University.
The subtilisin-like proteinase (AprBp) of Bacillus pumilus 3-19 is a potential candidate for industrial use as a feed additive. But to acquire a greater yield of the enzyme, it is important to create an extremely effective expression system.
The current research intends to acquire stable expression of the B. pumilus 3-19 protease gene in the Pichia pastoris expression system and characterize the relation of enzyme activity with the choice of vector type and signal peptides.
In the current research, the vectors pPINK-HC and pPINK-LC were chosen for the stable expression of the optimized protease gene in methylotrophic yeast cells, and a set of effective signal peptides were employed for extracellular secretion of recombinant protease.
The presequence of the α-mating factor repeatedly leads to a rise in the production of recombinant proteins. However, the sequences of killer and lysozyme signal peptides were not employed earlier for the expression of bacterial protease in P. pastoris.
Colonies of transformants containing constructs based on the pPINK-LC vector were identified on the third day of incubation and showcased greater transformation efficacy when compared to colonies with constructs based on the pPINK-HC vector identified only on the seventh day of incubation.
The transformation efficacy on the 10th day of incubation for structures based on the pPINK-LC vector averaged 1.3 ×103 transformants/μg DNA, while 1.03 × 103 transformant/μg DNA was documented for constructs with the pPINK-HC vector. Colony PCR revealed that all chosen transformant colonies had the subtilisin-like protease gene incorporated into the yeast genome.
The research revealed that the incubation time influences the production of protease in P. pastoris, and the maximum activity of the enzyme was noted.
Yeast strains containing constructs based on the low-copy vector pPINK-LC displayed greater protease activity (U/mL) in the hydrolysis of azocasein (2.49 ± 0.08 for α-mating factor presequence, 2.63 ± 0.16 for killer signal peptide (SP), 2.19 ± 0.11 for lysozyme SP).
The strains containing constructs based on the pPINK-HC vector displayed lower protease activity when compared to the above constructs (2.21 ± 0.07 for α-mating factor presequence, 1.86 ± 0.09 for killer SP, 1.31 ± 0.11 for lysozyme SP), irrespective of the signal peptide used.
The highest protease activity was obtained for yeast strains with pPINK-LC-killer-aprBp (5.75 ± 0.08 U/mL) and pPINK-LC-α-mat.factor-aprBp (4.33 ± 0.07 U/mL) constructs. The research revealed that the subtilisin-like protease from recombinant P. pastoris strains shows proteolytic activity, which relies on the incubation time and the choice of signal peptide and vector.
The generation of bacillary protease by the heterologous yeast-based expression system makes this system reassuring for the creation of novel feed additives for animal husbandry.
Proteases are placed among the three major groups of industrial enzymes and they account for around 60% of the total worldwide sale of enzymes, of which 40% belong to bacterial proteases. Bacterial serine proteases have broad substrate specificity and greater resistance to different conditions—which permits their use in biotechnology—specifically as bio-additives for farm animals and birds.
Moreover, feeds supplemented with proteases increase amino acid absorption by animals and save funds meant for buying synthetic amino acids. It is also revealed that using proteases as feed additives heightens the bioavailability of nitrogen, which alternately decreases the levels of ammonia pollution in the soil.
Furthermore, proteases promote the action of other enzymes and intensify the cost-efficiency of feeds, while also enhancing the growth of beneficial microflora in the gut of broilers.
At present, a limited number of commercial proteases from foreign manufacturers are used in Russia. The major representatives on the Russian market are the imported feed additives RONOZYME® ProAct (DSM (9.4%), Switzerland), CIBENZA DP100 (NOVUS (88.1%), USA), and Axtra® PRO TPT (DuPont (2.4%), Germany).
The recombinant obtained contains P. pastoris strains with subtilisin-like bacillary protease (aprBp) gene and three varied signal peptides (killer protein, α-mating factor, and lysozyme). Additional expression and purification of the protease from yeast culture liquid and the analysis of its biochemical and enzymatic properties would help characterize the biotechnological prospects of the recombinant enzyme.
Source:
Journal reference:
Pudova, D. S., et al. (2021) Heterologous Expression of Bacillus pumilus 3–19 Protease in Pichia pastoris and Its Potential Use as a Feed Additive in Poultry Farming. BioNanoScience. doi.org/10.1007/s12668-021-00899-2.