In this interview, we speak to Dr. Matthew Baker about his latest research that led to the programming of DNA 'robots' that are able to prod cell membranes.
Please can you introduce yourself and tell us about what inspired your latest research into DNA robots?
My work has focused on molecular machines, in particular the large molecular complex that makes bacteria swim, for some time. We have used this complex machine to try and understand the basis of emergent complexity, how you put simple things together which can then have a more complex overall function.
One way to attack this problem is using bottom-up synthetic biology, and so to that end, I also do work trying to build up more and more complex systems using simple parts. In this case, that is using simple bits of DNA to connect synthetic droplets to work towards more complicated droplet processing.
Can you describe how you carried out your latest research into DNA robots? How were these ‘robots’ built?
I think perhaps ‘robot’ is a bit of a fancy word to use here as it implies something quite complex and does a lot of stuff. Plenty of DNA nanotechnology is like this, complex and precise 3D structures that can change shape to ‘do’ something.
However what we did here was one step more basic than that, it was really just using 2-3 strands of DNA in a small complex to try and tie two droplets together, or prevent them from joining together, and most of this work was actually about how best to get DNA and these lipid droplets to work together. So not really ‘robots’, but a simple ‘string’ of maybe 4-5 DNA strands which could be untied with an extra piece of DNA to unravel the string and let the two species separate.
How is your DNA nanotechnology able to ‘punch holes’ in cell membranes? What applications can this have within science and health?
This is ultimately why we were doing all of this though because others have made larger and bigger pores made out of DNA (including ourselves) and we wanted to understand how best to decorate these pores with cholesterols so that they would go into the membrane as we intended.
At the end of the day the cholesterol group can be decorated around the DNA structure and this then stabilizes the normally water-loving DNA to be able to sit in the inside of the membrane, which is hydrophobic.
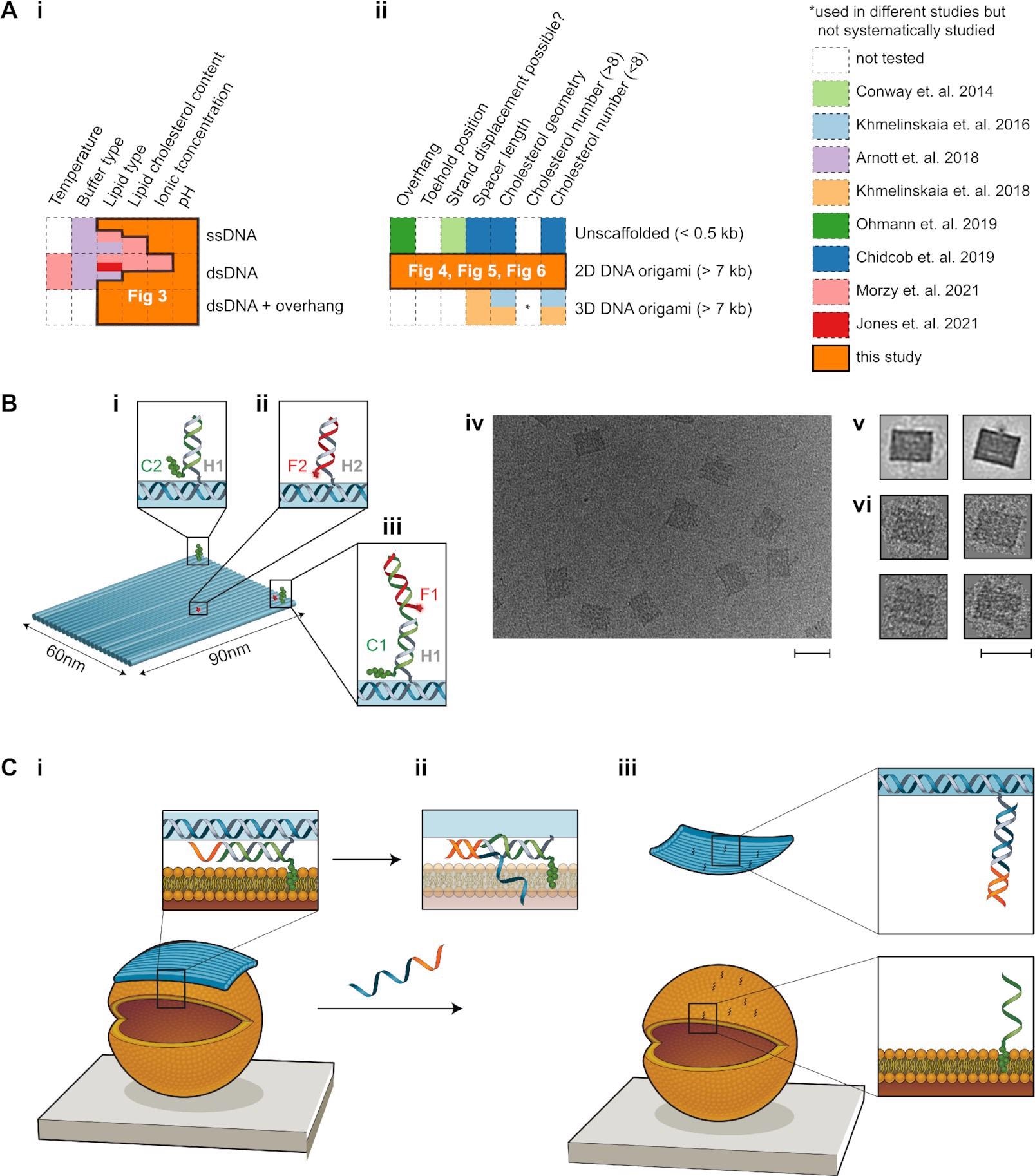
Image Credit: Binding of DNA origami to lipids: maximizing yield and switching via strand displacement
How will your research have implications within biosensing and mRNA vaccines?
We are building the essential basic technology for future droplet computation. For example, much like the transistor enables ultimately computers, here we are building simple DNA-based technology that will allow droplets to process signals.
For example, you might have some protein-based light-sensing technology in one droplet that then responds to a signal and changes the pH in that droplet, which then results in one of our DNA-based devices punching into a membrane and transmitting a signal to an adjacent droplet. That would be an example of a light-based sensor, and we can scale up this complexity with more sensors, and more complicated logic gates, to build a water-soluble, human-compatible sensor that flows around, say, your body, and records information (in DNA) to be read out later.
Previously, scientists have struggled to find the right buffer conditions for these lipids and liposomes. Why was this and how did you overcome this problem?
There hadn’t previously been a comprehensive exploration of buffer conditions, as well as in the different ways you can arrange the cholesterol group. This is primarily because I think people were happy with systems that worked rather than finding the ‘best’ route, but also because most people were coming from either a DNA background or a lipid background.
In our case, Dr. Shelley Wickham brings a lot of expertise with DNA, and I (supposedly) supply some experience with lipids, so we were able to optimize for both at once.
Liposome nanotechnology is one area of science that has shot into the limelight due to the use of liposomes alongside RNA within COVID-19 vaccines. Do you believe that as different methodologies are being highlighted due to the ongoing pandemic, researchers will start to trial new ways of doing research leading to more scientific advancements?
Certainly RNA work, and liposomes, have both had a huge increase in their popular awareness during the pandemic! I think there has never before in history been anything like the unified global endeavor to combat COVID-19, both through structural biology, as well as liposome and chemical biology and RNA.
The issue was simply so important that many different areas came together to focus on a goal. I am not sure if the methods of research have changed, rather than the immediate focus, and the urgency of the problem perhaps allowing new solutions to be found more quickly.
Drug delivery is one area of research that has faced struggles in past years with new innovative ideas being urgently needed. How can your new technology help to improve drug delivery systems?
The advantage of DNA-based systems that are built from the bottom up is that they allow us to control where everything goes and how it is put together. So we can design specific triggers to open a cage or respond to an environment. However the actual encapsulation, movement, and delivery of cargo are still very hard problems.
What DNA nanotechnology offers, in conjunction with liposome work, is more tunable interactions, and at one level, total control over the structure and kinetics of the de novo structures we build.
Image Credit: luminance studio/Shutterstock.com
What are the next steps for you and your research?
We are now trying to use our learnings about how best to get DNA and liposomes to work together to do more functional things with more complex DNA structures, such as building new types of pores and also sending specific signals between droplets to demonstrate controlled signal processing.
Where can readers find more information?
About Dr. Matthew Baker
Dr. Matt Baker is a Scientia Senior Lecturer in the School of Biotechnology and Biomolecular Sciences at the University of New South Wales.
Matt completed his DPhil in Physics at Oxford University as a John Monash Scholar studying the bacterial flagellar motor that makes nearly all bacteria swim. Subsequently, Matt investigated protein transport in Oxford's Chemistry Research Laboratory and the Department of Biochemistry. Upon his return to Australia, Matt focussed primarily on how simple subunit interactions govern assembly of complex architectures, including the rotor (NSMB 2016) and filament (eLife 2017). The next question is how this complexity emerged. To begin addressing this, Matt's nascent group at UNSW looks at how ion selectivity changes using directed evolution (Mol Micro 2019, Front. Microbiol. 2020) to examine the evolutionary landscape that constrains the adaptation of the motor (bioRxiv 2021).
Our group also investigates force sensitive proteins that in synthetic lipid bilayers (Channels 2019). These are essential at all scales of life, in bacteria to respond to osmotic shock and in mammals in the sense of touch and hearing and are involved in cancer progression and arthritis. In collaboration with Dr. Shelley Wickham, we probe membrane dynamics and interactions using novel DNA nanotechnology (Nanoscale 2019, NAR 2021), with the ultimate goal to control membrane communication using de novo DNA nanotechnology.
Matt also has a love of radio: he was a Top 5 Under 40 Scientist in Residence at the ABC in 2015 and has continued since then as the regular science presenter on Nightlife, broadcast on ABC Local across Australia, and by producing content for the Health Report, Science Show, Saturday Extra and Earshot on ABC's Radio National. He is currently a 'Resident Scientist' on Koori FM - Sydney's First Nations Radio station and also on Radio New Zealand's flagship weekend breakfast show Saturday Morning with Kim Hill. He also has applied his data skills to journalism; he was Australia's inaugural Google Newslab Fellow in 2016 when he was embedded at the Sydney Morning Herald. Matt is active in Australia-China relations and has been an delegate and panel chair at the Australia-China Youth Dialogue since 2015.
Currently, our group consists of 2 Ph.D. students, 3 postdocs, and 2 Hons students working in roughly a fifty-fifty split across flagellar and in vitro lipid/DNA projects. At any stage we welcome applications for PhDs and Hons students. We have recently been fortunate to be funded by the Human Frontier Science Program, so applicants interested in postdoctoral opportunities should get in touch.
A collection of links can be found on Matt's linktree.
Matt teaches in the School of Biotechnology and Biomolecular Science. He is co-convener of 3rd Year synthetic biology (BABS3200) focusing on DNA nanotechnology and directed evolution approaches, and he gives guest lectures into 1st Year Genes, 2nd Year Microbiology, 3rd Year Environmental Microbiology and 3rd Year Microbial Genetics.