Reviewed by Danielle Ellis, B.Sc.May 31 2023
Many of life’s secrets are hidden in the initial stages of embryonic development. Unraveling these mysteries can aid in the understanding of early development and birth abnormalities, as well as the creation of innovative regenerative medicine treatments.
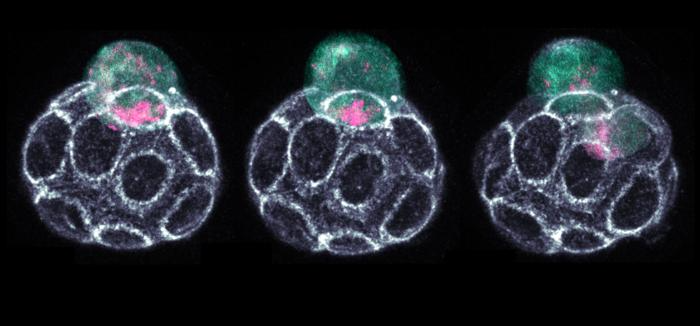 Living mouse embryo transitioning from 16- to 32-cell stage, showing the unequal inheritance of RNA (pink) and endoplasmic reticulum (cyan) to inner and outer daughter cells respectively. Image Credit: Azelle Hawdon.
Living mouse embryo transitioning from 16- to 32-cell stage, showing the unequal inheritance of RNA (pink) and endoplasmic reticulum (cyan) to inner and outer daughter cells respectively. Image Credit: Azelle Hawdon.
Researchers from the Australian Regenerative Medicine Institute (ARMI) at Monash University have categorized a pivotal phase in mammalian embryonic development using novel and robust imaging techniques. The study was published in Nature Communications.
Just a few days into the journey of embryogenesis, when turning into 16 cells, the embryo must make its first difficult decision - which of its cells will give rise to the embryo or will become extra-embryonic tissue, for example, placenta.”
Dr Jennifer Zenker, Lead Researcher, Monash University
The research group revealed how this decision-making process is aided by capturing the inner organization of single cells in the early embryo in this study.
Ribonucleic acid, RNA, plays a key role here. At the 16-cell stage, the different subtypes of RNA, named rRNAs, mRNAs, and tRNAs, are sorted to the two ends of a cell called apical and basal side. The distribution of RNA subtypes determines what the next generation of cells of the embryo will become.”
Dr Jennifer Zenker, Lead Researcher, Monash University
Surprisingly, while most mRNAs and tRNAs stay on the apical side, most rRNA molecules hitchhike down to the basal side on organelles called lysosomes. Despite having a lower total RNA content, the apical sides of outer 16-cell stage cells have all of the RNAs and other components essential for protein production.
The crowded basal side, on the other hand, is dominated by rRNAs. Daughter cells that acquire the apical side’s more active protein factories are more transformable and specialize in the future placenta.
Image Credit: Adao/Shutterstock.com
The daughter cells with pluripotency, or the ability to become any type of cell in the adult organism, acquire the less translationally active bulk of rRNA.
This and similar decisions, known as cell fate, are vital in development because they affect how these early cells evolve into their final cell type, like skin cells, heart muscle cells, and brain cells. Being able to orchestrate cell fate opens up the possibility of developing new stem cell-based treatments for a variety of diseases and conditions in regenerative medicine.
As in real life, cells can influence the direction of their own future by getting organized early. Our research may open new ways to predict and direct cell fate decisions.”
Dr Jennifer Zenker, Lead Researcher, Monash University
Source:
Journal reference:
Hawdon, A., et al. (2023). Apicobasal RNA asymmetries regulate cell fate in the early mouse embryo. Nature Communications. doi.org/10.1038/s41467-023-38436-2.