Mass spectrometry is a sophisticated technology that allows scientists to break down and identify the constituents of almost anything by measuring the mass of the microscopic particles that make it up. It has a significant restriction, however: around 99% of the material being examined is often lost before analysis begins.
 The new nanotube also has the unique ability to transfer ions that are dissolved in water directly into the vacuum of mass spectrometers. Image Credit: Nicholas Drachman.
The new nanotube also has the unique ability to transfer ions that are dissolved in water directly into the vacuum of mass spectrometers. Image Credit: Nicholas Drachman.
This rate of loss reduces the technology's potential. It decreases accuracy and sensitivity, consumes resources, and complicates sample preparation, perhaps leading to more mistakes. However, this could not last much longer. Brown University researchers have discovered a novel way for transporting the ions that mass spectrometers monitor, substantially minimizing sample loss and ensuring that virtually all of it remains intact.
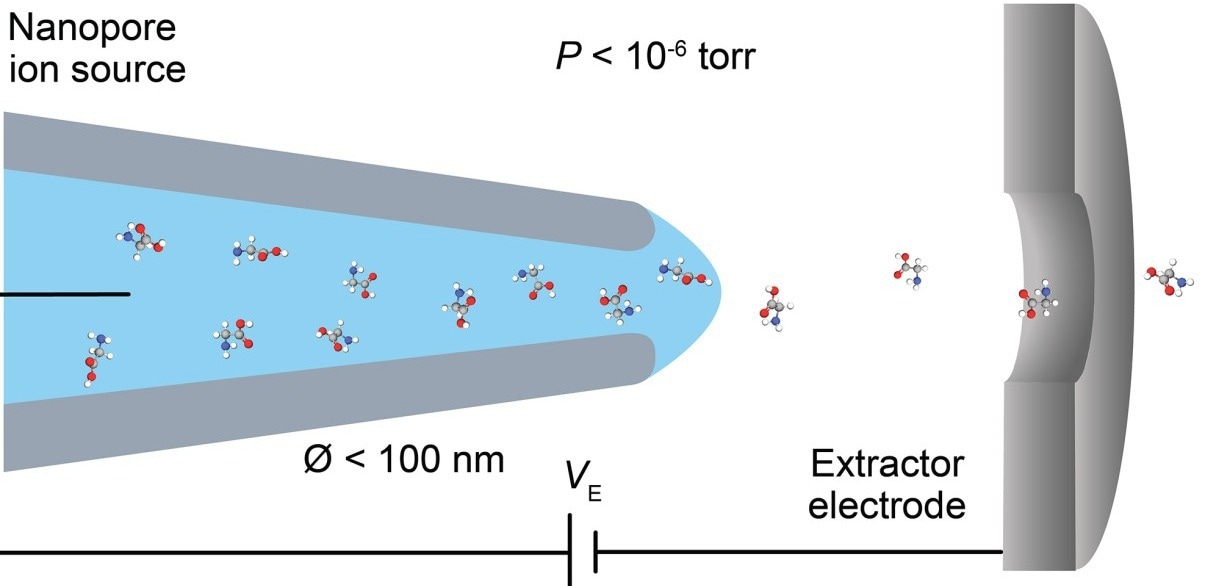
The conventional technique for producing ions for mass spectrometry, called electrospray ionization, basically involves a very sharp needle getting placed just in front of the mass spectrometer, hitting it with an electric field that pulls out a spray of charged droplets that eventually dry out to produce bare ions that make it into the mass spectrometer from open air.”
Nicholas Drachman, PhD Student, Brown University
Drachman added, “Basically, it is a process where you are really spraying your sample all over the place to produce these ions and only get a tiny portion of them into the mass spectrometer’s vacuum for analysis. Our approach skips all of that.”
The breakthrough, known as a nanopore ion source, breaks down a long-standing scientific barrier and has the potential to transform mass spectrometry technology. The Brown team presents their innovative invention in Nature Communications.
The key is a tiny capillary produced by the researchers, with an aperture of around 30 nanometers — nearly 1,000 times smaller than the width of a human hair. In contrast, the normal electrospray needle has an aperture of around 20 micrometers, which is over 600 times larger than the tube produced at Brown.
The novel nanotube also has the unusual capacity to transmit ions dissolved in water straight into the vacuum of a mass spectrometer, as opposed to creating a spray of droplets that must be dried before accessing the ions.
Furthermore, conventional mass spectrometers often suck in a large volume of gas along with the ions throughout the operation, needing many stages of vacuum pumping to pull in the ions. According to the experts, the new discovery eliminates the need to pump gas out because it will not be sucked in.
Drachman added, “Rather than place it in front of a mass spectrometer and generate this spray of droplets, we just place it directly into the mass spectrometer skipping this messy spray, drying and vacuum process. By generating ions in the vacuum directly, it drastically reduces the pumping requirements, which should significantly simplify the complex hardware of mass spectrometers.”
The Brown team was inspired by nanopore sequencing in DNA and intends to commercialize its concept for general usage by protein researchers. This includes the long-awaited objective of sequencing proteins one amino acid at a time.
Mass spectrometry is the best way to look at proteins, which are made up of amino acids that have all sorts of different chemical and physical properties, because it can tell them apart by the mass of their ions with high certainty. Proteomics have not seen the same advances as genomics in the last two decades, and so there is been this this hunger for a technology that can improve analysis of proteins.”
Derek Stein, Study Lead Author and Professor, Brown University
Stein added “By getting rid of that sample loss problem, it should enable these much more sensitive analyses to be possible, like sequencing the amino acids in a protein molecule one-by-one and in sequential order. This is the blue-sky idea that has motivated our work.”
The team spent the last ten years developing the new approach. They began by custom constructing their own mass spectrometer that could house the unique ion source in a vacuum, as opposed to standard designs in which the ion source is separate from the instrument and sits in the open air.
The researchers created the crucial component of its transfer mechanism by heating a glass tube in the center and then carefully pulling it apart to produce an incredibly small aperture at the tip that is undetectable to the human eye.
Trial and error played an important role in the process, frequently resulting in weeks of frustration as they sought to get everything working consistently at the tip of the capillary, which is far too small to check by eye.
Stein stated, “There were some weeks we did not know if we were cursed by God himself or something things just stopped working. Other weeks, everything worked brilliantly.”
The team's tenacity paid off. They successfully proved that ion analysis with their new transfer approach matches detections made with standard methods but with far reduced sample loss, providing a more efficient and accurate way to investigate microscopic particles.
“We needed to convince people in the proteomics field that we can generate the same kind of ions that they are used to generating by conventional electrospray and that we can do it in this different and, we believe, better way,” Drachman stated.
The study's analysis provides proof of concept for the approach. The researchers' next goal is to maximize the performance of their nanopore ion source.
Drachman concluded, “We need to show that this can improve the workflow of proteomic analyses. We would like to take that to the next level and make it something that will improve the science of researchers throughout the field.”
Source:
Journal reference:
Drachman, N., et al. (2024) Nanopore ion sources deliver individual ions of amino acids and peptides directly into high vacuum. Nature Communications. doi.org/10.1038/s41467-024-51455-x.