Reviewed by Lauren HardakerSep 3 2025
In a recent Cell Biomaterials study, University of Pennsylvania researchers offer AMP-Diffusion, a generative AI tool for creating tens of thousands of novel antimicrobial peptides (AMPs), short strings of amino acids that serve as the building blocks of proteins and have bacteria-killing capability. In animal models, the most powerful AMPs performed similarly to FDA-approved drugs, with no noticeable side effects.
 Image credit: Christoph Burgstedt/Shutterstock.com
Image credit: Christoph Burgstedt/Shutterstock.com
What if generative AI could create life-saving antibiotics, rather than merely art and text?
While previous achievements at Penn have demonstrated that AI can effectively search through mounds of data to uncover viable antibiotic possibilities, this work adds to a tiny but increasing body of evidence that AI can create antibiotic compounds from scratch.
Nature’s dataset is finite; with AI, we can design antibiotics evolution has never tried.
César de la Fuente, Study Senior Co-Author and Presidential Associate Professor, School of Arts & Sciences, University of Pennsylvania
“We’re leveraging the same AI algorithms that generate images, but augmenting them to design potent new molecules,” added Pranam Chatterjee, Assistant Professor and Study Other Senior Co-Author, BE and Computer and Information Science, Penn Engineering, who began work on the project while at Duke University.
Two Labs, One Goal
For years, de la Fuente's group has successfully used artificial intelligence to seek molecules with antimicrobial capabilities in odd locations, including woolly mammoth proteins, animal venom, and ancient bacteria known as archaea.
“Unfortunately, antibiotic resistance keeps increasing faster than we can discover new antibiotic candidates,” said de la Fuente.
As a result, his lab collaborated with Chatterjee's, which generally uses AI to build peptides to cure diseases when traditional drug development approaches have failed.
“It seemed like a natural fit. Our lab knows how to design new molecules using AI, and the de la Fuente Lab knows how to identify strong antibiotic candidates using AI,” added Chatterjee.
 Study: Generative latent diffusion language modeling yields anti-infective synthetic peptides
Study: Generative latent diffusion language modeling yields anti-infective synthetic peptides
Tuning Out the Noise
While some generative AI models, like ChatGPT, work by anticipating the next word or element in a sequence, “diffusion” models start with random “noise” and repeatedly refine it into a coherent output. This idea is behind tools like DALL·E and Stable Diffusion.
AMP-Diffusion operates similarly, except it refines amino acid sequences instead of "denoising" pixels.
“It’s almost like adjusting the radio. You start with static, and then eventually the melody emerges,” said de la Fuente.
At least two other research teams have used diffusion models to create antimicrobial peptides, but AMP-Diffusion takes a unique method.
Instead of initially building its own protein "latent space" — an internal map of how proteins are formed — AMP-Diffusion relies on Meta's ESM-2, a widely used protein language model based on hundreds of millions of real protein sequences.
Since ESM-2 already has a detailed "mental map" of how genuine proteins go together, AMP-Diffusion does not need to re-learn fundamental biology. This implies it can create candidate AMPs faster, and its outputs are more likely to follow the complicated patterns that peptides use.
Chatterjee’s team also created AMP-Diffusion to consult ESM-2's built-in guidelines when “denoising,” thereby providing the new tool a coach to keep it anchored in biological reality.
“Instead of teaching the model the ABCs of biology, we started with a fluent speaker, that shortcut lets us focus on designing peptides with a real shot at becoming drugs,” said Chatterjee.
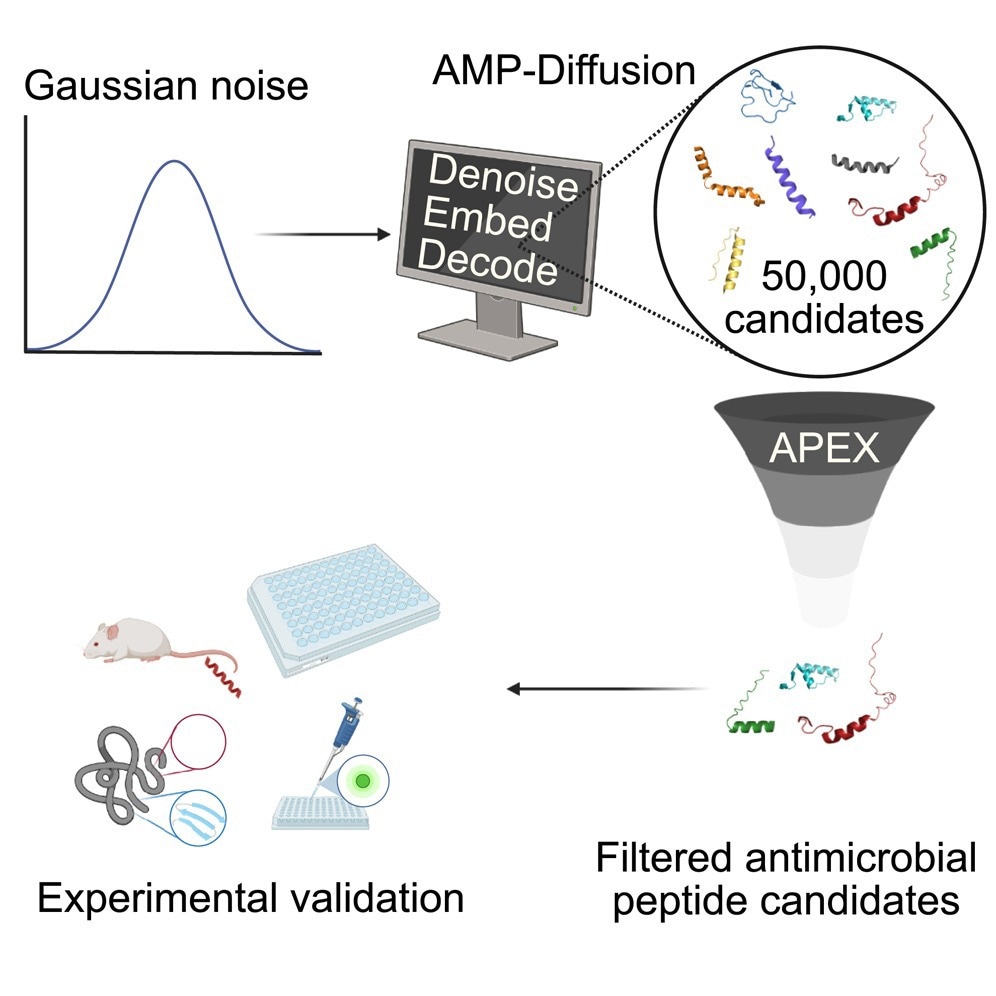
From 50,000 Designs to Two in vivo Winners
Using AMP-Diffusion, the researchers synthesized amino acid sequences for over 50,000 candidates.
“That’s far more candidate drugs than we could ever test. So we used AI to filter the results,” stated de la Fuente.
APEX 1.1, an AI tool built by de la Fuente's group, was fine-tuned by searching for antibiotic possibilities ranging from ancient microbial proteins to Neanderthal proteins. It graded the prospective AMPs based on a variety of parameters. These included estimating which sequences would be effective bacteria-killers, removing peptides that were too close to known AMPs, and ensuring that the remaining candidates spanned a wide variety of sequence types.
After developing the 46 most promising possibilities, the de la Fuente lab evaluated them in human cells and animal models. Two AMPs treated skin infections in mice with the same efficacy as levofloxacin and polymyxin B, FDA-approved medications used to treat antibiotic-resistant bacteria, and had no side effects.
“It’s exciting to see that our AI-generated molecules actually worked. This shows that generative AI can help combat antibiotic resistance,” added Chatterjee.
Next Steps for AI-Generated Antibiotics
In the future, the researchers intend to improve AMP-Diffusion, allowing it to optimize for a more precise purpose, such as treating a particular type of bacterial infection.
“We have shown the model works, and now if we can steer it to enhance beneficial drug-like properties, we can make ready-to-go therapeutics,” noted Chatterjee.
The researchers' latest study is proof of principle: generative AI can go beyond mining what evolution has previously built and develop new drugs.
“Ultimately, our goal is to compress the antibiotic discovery timeline from years to days,” concluded de la Fuente.
Source:
Journal reference:
Wan, F., et al. (2025) Generative latent diffusion language modeling yields anti-infective synthetic peptides. Cell Biomaterials. doi.org/10.1016/j.celbio.2025.100183