 By Pooja Toshniwal PahariaReviewed by Lauren HardakerSep 10 2025
By Pooja Toshniwal PahariaReviewed by Lauren HardakerSep 10 2025In a study published in Metabolic Engineering, researchers engineered Escherichia coli to biosynthesize 2,5-pyridinedicarboxylate (2,5-PDCA) from glucose via p-aminobenzoic acid (PABA). The team achieved a record titer of 10.6 g/L, surpassing the previous microbial benchmark, offering a promising route toward bio-based alternatives to fossil-derived plastic precursors.

Image credit: YRABOTA/Shutterstock.com
The breakthrough offers a potentially scalable and renewable alternative to fossil fuel–based plastic precursors, though the need for pyruvate supplementation remains a barrier to immediate industrial applications. The work highlights microbial engineering as a powerful route to sustainable materials, aligning with global efforts to reduce greenhouse gas emissions and advance the circular bioeconomy.
Plastics derived from fossil fuels significantly contribute to greenhouse gas emissions, creating urgent demand for renewable, sustainable alternatives. Bio-based biodegradable plastics offer a promising solution, and PDCA, particularly 2,5-PDCA, has gained attention as a substitute for terephthalic acid in polyethylene terephthalate (PET). Previous polymer studies have shown that its derivatives enable polymers with enhanced strength, stability, and polymerization.
However, chemical synthesis of 2,5-PDCA suffers from poor selectivity and low yield, which limits its potential for industrial application. Microbial production from biomass represents a sustainable alternative; however, efficient and scalable pathways remain underdeveloped, making it crucial to develop innovative biotechnological strategies to enable large-scale, eco-friendly plastic production.
About The Study
In the present study, researchers established a biosynthetic pathway for microbial production of 2,5-pyridinedicarboxylate (2,5-PDCA) using glucose and engineered Escherichia coli. They designed a two-module strategy involving PABA biosynthesis and 2,5-PDCA conversion. While previous studies report individual reactions, this work represents the first integrated de novo pathway enabling direct glucose-to-2,5-PDCA conversion.
Pathway design was guided by flux balance analysis (FBA) using the E. coli metabolic model iML1515 with heterologous reactions added for 2,5-PDCA biosynthesis via PABA. Codon-optimized genes, protocatechuate 2,3-dioxygenase (praA, from Paenibacillus), 4-amino-3-hydroxybenzoate 2,3-dioxygenase (ahdA, from Bordetella), and p-hydroxybenzoate hydroxylase (pobA, from Pseudomonas aeruginosa and Caulobacter vibrioides) from bacterial sources, were synthesized and cloned using polymerase chain reaction (PCR), overlap PCR, and plasmid construction.
Sequentially introducing the PABA module (pabABC) and the 2,5-PDCA module (CvpobA and ahdA) into host strains CFT1 and CFT5 generated recombinant Escherichia coli strains. The latter carried gene deletions (tyrA, pheA, pykA, pykF) to redirect flux toward shikimate and PABA biosynthesis.
The team performed cultivation in lysogeny broths (LB) and M9Y mediums, supplemented with glucose (15–30 grams/liter), antibiotics, and sodium pyruvate as indicated. Shake-flask cultures were initiated at an optical density of 0.05–0.2 at 600 nanometers (OD600) and incubated at 30–37°C with shaking at 220 rpm. Fed-batch fermentation was conducted in a 1-L bioreactor under controlled pH (7.0), temperature (30°C), and dissolved oxygen (DO) conditions.
To mitigate oxidative stress on the PABA-converting enzyme (AhdA), pyruvate supplementation and the expression of oxidative stress resistance genes (katE, katG, ytfK) were employed. DO was optimized to minimize excess oxygen, and fed-batch feeding was initiated once glucose levels fell below 5.0 g/L.
Analytical methods included OD600 for growth, high-performance liquid chromatography (HPLC) for metabolite quantification, and quantitative reverse transcription PCR (qRT-PCR) for transcriptional analysis. Hydrogen peroxide (H₂O₂) tolerance assays indicated resistance to oxidative stress. The researchers measured final 2,5-PDCA titers following anion-exchange purification and freeze-drying of culture supernatants.
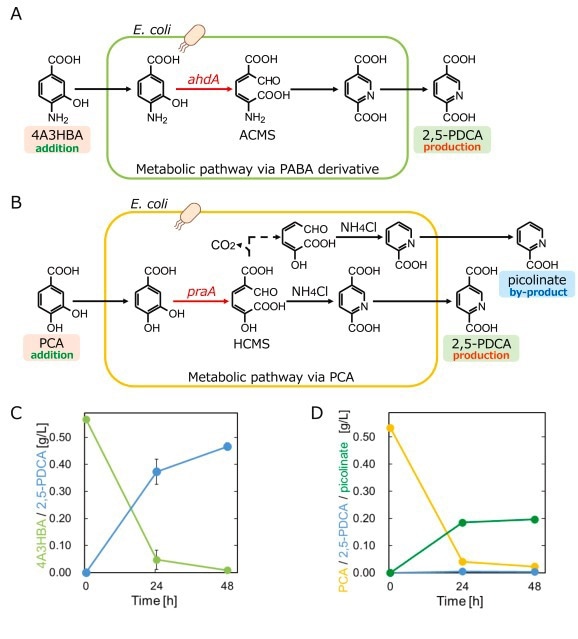
Production of 2,5-pyridinedicarboxylate from the precursor protocatechuate or 4-amino-3-hydroxybenzoate Schematics illustrating 2,5-PDCA production (A) from 4A3HBA and (B) from PCA. (C) Production of 2,5-PDCA (light blue circles) from 4A3HBA (light green circles) in CFT1-ahdA. (D) Production of 2,5-PDCA (light blue circles) and picolinate (green circles) from PCA (orange circles) in CFT1-praA. Data are presented as mean values from three independent experiments, with error bars indicating standard deviation.
Results
The engineered E. coli strains successfully converted glucose molecules into 2,5-PDCA through a modular pathway integrating PABA biosynthesis and conversion. Under optimized conditions, the process produced 1.8 g/L of 2,5-PDCA in test-tube cultures after 72 hours. Bioreactor fermentation further improved performance, generating almost 11 g/L after 144 hours (at OD600 of 60), the highest 2,5-PDCA produced through microbial fermentation.
Minimal PABA accumulation and the absence of detectable 4-amino-3-hydroxybenzoate (4A3HBA) levels throughout the cultivation period indicate efficient metabolic channeling. Expression of ahdA converted 0.5 g/L of 4A3HBA into 0.47 g/L of 2,5-PDCA (76.5% efficiency), whereas the PCA route produced only 0.04 g/L, confirming PABA superiority. Among pobA variants, CvpobA showed the most effective hydroxylation, generating 0.47 g/L of 4A3HBA and supporting efficient module assembly.
Oxidative injury to the PABA-transforming enzyme, probably caused by H₂O₂ released during the process, was overcome by supplementing with pyruvate, which alleviated oxidative inactivation of PobA. Decreasing pyruvate supplementation from 3.0 g/L to 1.0 g/L also generated 8.0 g/L, indicating potential for cost-effective industrial application.
Unlike previously documented pathways of pyridine acid synthesis, which incorporate nitrogen through chemical or non-biological mechanisms, the study highlights the promising potential of enzyme-catalyzed nitrogen incorporation as an effective and sustainable strategy. This approach opens avenues for advancing the biological manufacturing of nitrogen-comprising aromatic compounds.
Conclusions
The study demonstrates an efficient microbial platform for producing 2,5-PDCA from glucose in E. coli, achieving the highest reported titer to date (10.6 g/L) through pathway integration, competing-pathway deletions, and pyruvate optimization. Enzymatic nitrogen incorporation provided a sustainable alternative to chemical methods, broadening opportunities for nitrogen-containing aromatics. Although pyruvate supplementation poses scalability challenges, reduced supplementation still yielded 8.0 g/L, indicating industrial promise.
Future directions include dynamic regulation of CvpobA, fine-tuning pyruvate kinase activity, balancing stress tolerance with metabolic burden to optimize scalability, and expanding this strategy to other pyridine carboxylates. These advances could enable cost-effective and large-scale bio-production of plastics and strengthen sustainable biomanufacturing platforms.
Download your PDF copy now!
Journal Reference
Katano, A., Mori, A., Nonaka, D., Mori, Y., Noda, S., & Tanaka, T. (2025). Biosynthesis of 2,5-pyridinedicarboxylate from glucose via p-aminobenzoic acid in Escherichia coli. Metabolic Engineering, 92, 252-261. DOI: 10.1016/j.ymben.2025.08.011. https://www.sciencedirect.com/science/article/pii/S1096717625001302?via%3Dihub