Researchers from the Institute for Genome Sciences at University of Maryland School of Medicine are aiming to perform high-throughput culturomics analysis of the vaginal microbiome, with the overall aim of developing live biotherapeutics capable of modulating the function and composition of the vaginal microbiome, improving women’s gynecological and obstetric health.
However, to achieve this goal, a better understanding of the intra-species diversity of the beneficial bacterium, Lactobacillus crispatus, is needed, as well as the performance of in-depth within-sample cultivation to obtain co-resident L. crispatus strains.
The traditional method for culturing of plating and colony picking has proven to be rather time-consuming and laborious.
As such, the team are developing methods for high-throughput culturomics analyses that integrate single-cell dispensing, bacterial sorting, and dilution-to-extinction culture. Through the dispensation of single cells into 384-well plates, hundreds or even thousands of single strains can be isolated in a single culture event.
Plated agar is utilized to confirm that single-cell colony-forming units are being dispensed. The dispensation of single cells into high-density wells ensures no cross-contamination of colonies, allowing robotics to re-suspend the colonies for greater storage and culture and increase the cultivation throughput.
For this process, 384-well plates have to be filled with agar medium. However, the dispensing of molten agar is limited by its viscosity (10-100 cps, 1.5% sol, 60 ˚C) and a small dispensing temperature range (~50-60 ˚C). Manual pipetting or pouring is also quite tedious and often results in bubble formation, making it difficult to discern colonies.
This study demonstrates the use of SPT Labtech's dragonfly® discovery for fast, small-volume, molten agar dispensing into 384-well assay plates followed by single-cell culture inoculation.

Image Credit: SPT Labtech
Method
The agar medium was prepared by adding agar (RPI, #9002-18-0) to Difco™Lactobacilli MRS Broth (BD, #288110) to a 1.5% w/v solution and autoclaved at 121 ˚C for 15 minutes, following the instructions of the manufacturer.
The bottle containing media was then relocated into a 65 ˚C water bath near dragonfly discovery and left to equilibrate before dispensing.
Using the dragonfly discovery software, a method was written for dispensing molten MRS agar utilizing the “Corning 384 Low volume flat bottom (24x16)” plate file in a snaking pattern with ultra-low retention (ULR) syringes (SPT Labtech, #4150-07208). The Dispense settings were altered to the “Gentle bulk addition above 5 μL” preset, to arrange a dispense speed of 3 mm/second.
Configurations for dispensing volumes and the number of syringes were optimized to ensure efficient syringe usage and to prevent agar from solidifying before the completion of the dispensing process.
To achieve a dispense of 40 µL across a single plate, four syringes were positioned at A2, A3, B2, and B3. Meanwhile, for dispensing 25 µL, six syringes were utilized at positions A2 to A4 and B2 to B4. This arrangement enabled the simultaneous dispensing across two plates using a multi-aspiration mode.
For the 40 µL and 25 µL protocols, the dispense times were 1 minute 45 seconds, and 35 seconds per plate, respectively. To maximize the time before agar solidification, the procedure involved engaging the syringes and adding standard reservoirs. Additionally, a 384-well assay plate (Corning, #3680) was positioned in the tray with its lid removed before initiating the instrument protocol.
Molten MRS agar was quickly transferred using a serological pipette, and an appropriate volume was added to each reservoir.
Once the last reservoir was filled, the protocol was immediately started. For each aspiration, the process was repeated with new syringes and reservoirs. Plates were allowed to cool in a laminar flow hood with the lid removed to prevent condensation.
The agar depths for the 40 µL and 25 µL dispense volumes were measured at 9.5 mm and 4.8 mm, respectively, as indicated in Table 1.
Table 1. Fill metrics and observed growth in 384-well agar filled plates. Source: SPT Labtech
| Dispense volume (μL) |
Agar depth (mm) |
% Growth wells |
Lambda (λ)* |
| 25 |
4.8 |
25.3 |
0.3391 |
| 40 |
9.5 |
39.7 |
0.5054 |
*Lambda (λ, taken from the Poisson Distribution formula) is equivalent to the plating density (number of viable cells/well) ƛ = -ln(growth negative wells/total wells) = average plating density (cells/well)
To test the 25 μL and 40 μL fill 384-well agar plates, dragonfly discovery was used to inoculate the agar-filled plates with single cells of L. crispatus from a culture diluted to extinction.
To plate at an average density of 0.5 bacteria/well, the stock culture was diluted in MRS broth to a density of 2 x103 CFU/mL. Dragonfly discovery was subsequently used to dispense 500 nL of culture to 368 wells of each plate, leaving column 24 uninoculated.
Plates were incubated anaerobically at 37 ˚C for 72 hours. Wells with colony growth were manually counted.

Image Credit: SPT Labtech
Results
With a plating density of 0.5 bacteria/well, the predicted percentage of growth wells was 39.3 % based on Poisson distribution. For the 25 μL and 40 μL fill plates, the proportion of positive wells was 25.3 % and 39.7 %, respectively (Table 1).
Colony growth was observed within the wells, covering the entire surface area of the agar, including the side walls and corners, as illustrated in Figure 1.
Multiple optical density readings of the plates, including well scans, were conducted in an attempt to identify positive wells. However, this approach yielded inconsistent results and was deemed unsuitable for data collection. The variability in results was likely attributed to the off-center positioning of colonies within the wells and the variation in agar depth away from the well center caused by the meniscus.
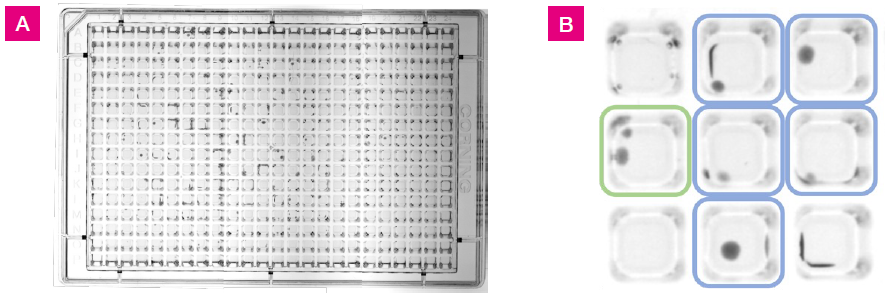
Figure 1. L. crispatus colony growth at 48 hrs on 384-well agar plate with 25 μL fill volume. A) Images of the plate were taken using a Syngene GBOX imaging system. This image was generated by stitching together three separate images. B) A section of the plate from wells G3 to I5. Colored squares indicate growth positive wells with one colony (blue) or two colonies (green). Colonies were located in the well centers and along the well walls. Image Credit: SPT Labtech
Conclusions
After considering the anticipated proportion of positive wells with growth, it was determined that the 40 µL fill volume was more suitable for the culture application. The 25 µL fill volume yielded approximately 36% fewer positive wells with growth than what was expected.
One possible hypothesis for this is that the trajectory of the inoculum dispense droplet may be angled in a way that causes the droplet to make contact with the side wall of the well rather than the agar. This scenario could be less likely in a 40 µL fill plate since the distance the droplet travels from the syringe is shorter.
About SPT Labtech
SPT Labtech designs and manufactures robust, reliable and easy-to-use solutions for life science. We enable life scientists through collaboration, deep application knowledge, and leading engineering to accelerate research and make a difference together. We offer a portfolio of products within sample management, liquid handling, and multiplexed detection that minimize assay volumes, reduce material handling costs and put the discovery tools back in the hands of the scientist.
At the Heart of What We Do
Many of our innovations have been born out of the desire to create solutions to existing customer problems; and it’s this ethos that drives SPT Labtech’s R&D efforts. Our strengths come from the trust our customers have with us to develop truly unique, automated technologies to meet their needs. We combine cutting edge science with first-rate engineering to put customers at the heart of everything we do.
A Problem-Solving State of Mind
The substantial breadth of expertise within our company enables us to be involved in the full life cycle of our products from the initial design concept, mechanical and software engineering and prototyping, to final manufacture and sale. These qualities allow us to offer the best possible technical and mechanical support to all the equipment that we supply, hence maintaining excellent client relationships.
Sponsored Content Policy: AZO Life Sciences publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of AZO Life Sciences which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.