Next-generation sequencing (NGS) has transformed the medical and biological sciences, delivering enhanced throughput and improved affordability.
The fundamentals of NGS library preparation remain mostly unchanged, despite a substantial drop in the time required (now taking less than half a day) and the cost of whole-genome sequencing (from $100 million in 2001 to just $200 in 2023).
NGS workflows demand accurate and precise liquid handling to satisfy sample preservation and high throughput demands.
This article discusses how NGS workflow can be automated and miniaturized with dynamic, low-volume liquid handling solutions to guarantee optimal sequencing depth and coverage for any application.
Know Your Methods: Sequencing Platforms and Library Preparation Approaches
Knowing the variety of sequencing modalities and library preparation methods can assist researchers in making the right experimental choices.
There are many common experimental considerations, such as throughput and sample type, genetic target size (e.g. a few genes or the whole genome), sample-to-result time, operational and experimental costs, and the sequence depth/coverage required.
Read length also influences the decision. While Sanger’s first-generation sequencing techniques are quick and economical for analysis of a small number of genomics targets, NGS can assess millions of short fragments at the same time, using a massively parallel sequencing approach.1
‘Bridge amplification’ is a second-generation sequencing technology promoted by Illumina. It is reliant on DNA synthesis to amplify and integrate modified fluorescent bases into short (50-500 bp) DNA fragments. These techniques deliver some of the highest levels of data accuracy and throughput.
Third-generation sequencing (long-read sequencing; LRS) delivers significantly longer readability (5-100 kb) and the opportunity to close gaps in knowledge within short-fragment assemblies.2
DNA fragments are individually sequenced as they are copied within individual microplate wells (Pacific Biosciences, referred to as PacBio) or as they move through protein nanopores (Oxford Nanopore Technology).
Despite higher error rates in long-read sequencing platforms than short-read, longer overlapping sequences provide a simpler assembly and enhance structural variant calling and full-length transcript reconstruction.
PacBio, Illumina, and Nanopore sequencing depend on a high-quality library input into the sequencer. The options for library preparation for NGS typically begin with genetic material.
DNA vs. RNA
DNA is priceless for the investigation of genetic conditions and for producing predictive assessments of a biological sample. However, it is not helpful regarding a cell’s dynamic, real-time operations.
In numerous disease states, the downstream expression of the encoded gene is what matters, not the DNA sequence itself. As a result, DNA sequencing (DNA-seq) and RNA sequencing (RNA-seq) may each be employed to answer different biological questions.
Bulk vs. Single-cell Analysis
The bulk analysis is useful if a sample consists of multiple, perhaps thousands of cells.
However, if detailed information about individual cells is required, though it is more costly and labor-intensive, single-cell analysis exposes cellular heterogeneity and expression profiles of cell subpopulations. Both are masked using bulk RNA sequencing.
Whole-genome vs. Whole-exome Sequencing
Whole-genome sequencing delivers the maximum resolution and enables the assembly of novel genomes. Conversely, whole-exome sequencing (WES) concerns only sequencing the protein-coding regions, approximately 3% of the human genome.
WES is the preferred method for clinical applications because of its cost-effectiveness, high coverage of regions of interest, a small amount of required raw data, and ease of analysis.
Identify Common Challenges: Data Reproducibility, Contamination, and Cost
NGS can be employed to overcome many biotechnological and clinical challenges associated with pathogen characterization and surveillance, drug development, cancer research, and diagnostics.3
Widespread concerns are present for all applications of NGS, such as the lack of reproducibility, scalability, contamination, and high reagent costs.
Reproducibility
The lack of reproducibility in research has been credited to numerous factors, with batch effects of experiments playing an essential part. The batch effect is the non-biological differences or technical variation between sample groups.
These complex artifacts can occur from many sources, including day-to-day sample processing, reagent batches, pipetting accuracy, reaction conditions, and even different technicians.
Automating liquid handling is a straightforward solution that guarantees consistency in genomics studies and enables samples to be processed in fewer batches.
Contamination
Sample-to-sample and environmental contamination frequently go undetected until the completion of data analysis. This wastes valuable bioinformatics and wet-lab time.4
Contamination can compromise data outcomes, especially in targeted sequencing workflows, as short-fragment amplicons can align to many species’ reference genomes. This means minimizing the risk of contamination from user error and environmental sources is fundamental.
Cost
Although sequencing costs per sample are lower than ever, NGS reagents are relatively costly. Utilizing lower reagent volumes per reaction is an effective way to enhance workflow affordability and economy.
Due to NGS reagent viscosities being variable (and frequently unknown to the researcher), selecting the correct liquid handling solution is vital for high-quality and reproducible NGS libraries, particularly at reduced volumes.
The careful design of experiments, excellent lab practices, and accurate liquid handling at each step of sample and library preparation (as displayed in Figure 1) all enhance the reliability of the sequenced data.
The introduction of contamination may occur at any stage. As a result, it is crucial to maintain accuracy and sterility during the whole process.
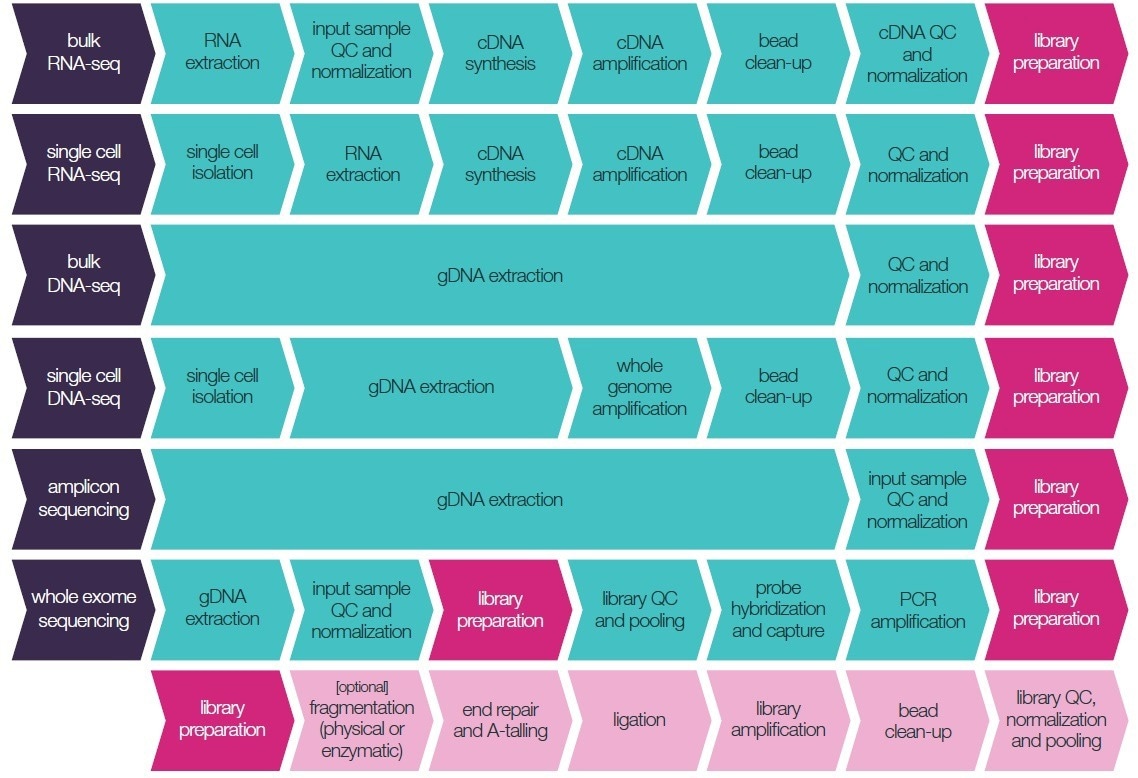
Figure 1. Typical NGS library preparation workflows available for miniaturization. Image Credit: SPT Labtech
Achieve Liquid Handling at a Fraction of the Volume
NGS miniaturization is especially valuable for high-throughput applications, such as drug discovery and the design and development of assays. It is all about finding a sweet spot between the quality of sequence data and a decrease in reaction volumes (and, consequently, experimental cost).
Different chemistries demand different reaction volume reductions. Some NGS kits are robust at a broad range of sample concentrations and volumes, whereas others enable a modest but impressive 4-fold volume reduction.
Sample type, such as bulk versus single-cell DNA/RNA, should also be considered, or low versus high RNA content.
In general, single cell enables significant miniaturization because the input is only a few picograms of genetic material. More complicated samples should be processed less aggressively to guarantee adequate depth and coverage of sequence data.
Aside from the decrease in experimental costs via the preservation of sample and reagent, advantages of NGS miniaturization include:
- Increased scope of applications – As researchers progressively push the boundaries of science, the capability to use lower volumes means that NGS could be applied to increasingly difficult types of samples.
- Higher-resolution libraries – Reducing reaction volumes while maintaining library quality, depth, and coverage can aid higher-resolution NGS libraries.
- Mitigation of batch effects – Decreasing reaction volumes enables researchers to process more samples from one kit and take a process from 96- to 384-well format, which further minimizes the batch effect.
- Increased throughput capacity – Savings in reagent volume deliver improved workflow economy and enhanced sample throughput. Automated liquid handling solutions support NGS miniaturization, which release technical staff to conduct more value-added tasks.
A broad range of sequencing protocols can be miniaturized with little-to-no expertise. NGS workflow miniaturization is now straightforward, affordable, and accessible, due to sophisticated, automated low-volume liquid handling technology.
Automation also provides further advantages for miniaturization workflows. For example, it supports reduced user variability and practically removes costly risks associated with reagent or sample contamination.
Choose a Dynamic Solution for NGS Workflow Miniaturization
SPT Labtech offers dynamic, automated liquid handling solutions that utilize both precise positive displacement pipetting and non-contact nanoliter-to-milliliter dispensing. As a result, they provide NGS miniaturization solutions over a wide variety of liquid types and applications.

mosquito® HV genomics. Image Credit: SPT Labtech
mosquito® LV genomics. Image Credit: SPT Labtech

dragonfly® discovery. Image Credit: SPT Labtech
The mosquito® genomics range of automated liquid handlers delivers highly precise multi-channel positive displacement pipetting, with the automation of pipetting of 500 nL to 5 µL volumes using the mosquito HV, or to greatly miniaturize at 25 nL to 1.2 μL volumes using the mosquito LV pipettor.
Using pre-sterilized disposable pipettes removes any risk of contamination, preserving data integrity without compromising efficiency or speed.

dragonfly® discovery. Image Credit: SPT Labtech
The dragonfly® discovery enables accurate and repeatable nanoliter-to-milliliter dispensing to be carried out, regardless of environmental conditions or liquid viscosity.
The instrument provides high-density plate format options (up to 1,536 wells) and employs powerful assay development software, making it the perfect platform for high-throughput genomics and maximum discovery power.
Employing disposable micropipette tips and positive displacement technology, SPT Labtech’s mosquito genomics liquid handlers effectively remove any environmental or sample-to-sample contamination risk.
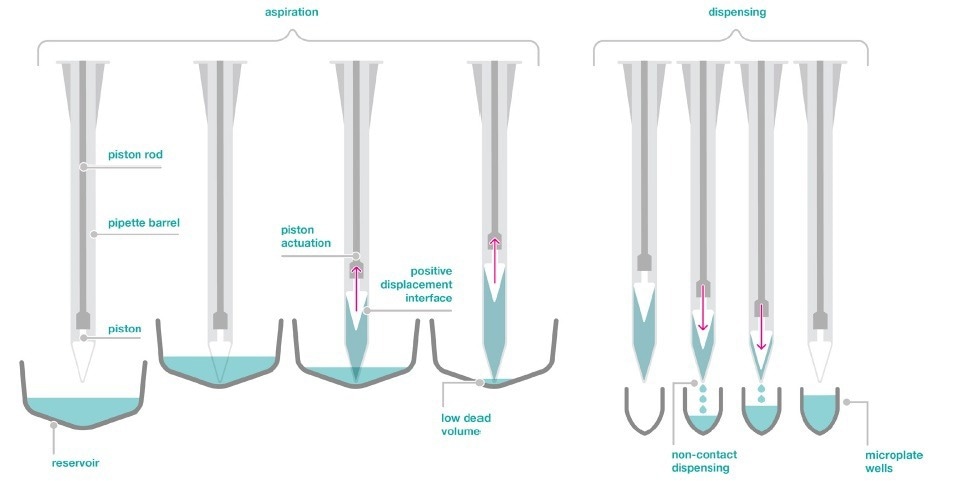
Figure 2. Positive displacement aspiration and non-contact dispensing technology of the dragonfly discovery. Image Credit: SPT Labtech
The dragonfly discovery also utilizes positive displacement to dispense in a non-contact way with high precision and accuracy, as shown in Figure 2.
Although each syringe is independent of the others, all syringes may be operated at the same time for rapid and highly flexible reagent distribution across a range of plate layouts.
The wide dispense range facilitates the miniaturization of NGS protocols and supports general liquid handling steps.
Choose from More than 40 Miniaturized Methods – or Create Your Own
SPT Labtech offers various automated liquid handling solutions that have assisted genomics researchers in successfully miniaturizing more than 40 protocols. Some examples include:
Affordable and Accurate RNA-sequencing
Miniaturized Illumina RNA-seq libraries were produced using the mosquito HV genomics pipettor. The libraries behaved like full-scale libraries, with comparable discovery power and reproducible sample clustering across cell types and treatments.5
Miniaturization was shown to optimize reagent usage and minimize the processing time per sample.
Single-cell and Single-nucleus Sequencing
Miniaturizing the Smart-Seq2 workflow enabled researchers to accomplish full-length, deep sequencing of single-cell and nuclear extracts.6
Utilizing the mosquito HV genomics with a 384-well plate setup, semi-automated methods that spanned reverse transcription, library preparation, and sample clean-up enabled the transcriptomic analysis of individual nuclei and each cell.
Long-read Sequencing
Miniaturized, automated approaches provide increased speed in the design and implementation of the protocol. This allowed long-read sequencing solutions to be developed in collaboration with PacBio, in response to the COVID-19 pandemic.7
The HiFiViral SARS-CoV-2 workflow employed the mosquito HV genomics pipettor to automate the high-throughput sequencing of SARS-CoV-2 from nasopharyngeal samples. Results exhibited more than 90% genomic coverage at 96- and 384-plex setups, with efficient variant calling for genomic surveillance.
You can learn more about improving the productivity of your genomics workflow here.
References and Further Reading
- Slatko BE, Gardner AF, Ausubel FM. Overview of next-generation sequencing technologies. Curr Protoc Mol Biol. 2018;122(1):e59. doi:10.1002/cpmb.59
- Rhoads A, Au KF. PacBio sequencing and its applications. Genom Proteom Bioinform. 2015;13(5):278-289. doi:10.1016/j.gpb.2015.08.002
- Yin Y, Butler C, Zhang Q. Challenges in the application of NGS in the clinical laboratory. Hum Immunol. 2021;82(11):812-819. doi:10.1016/j.humimm.2021.03.011
- Sangiovanni M, Granata I, Thind A. et al. From trash to treasure: detecting unexpected contamination in unmapped NGS data. BMC Bioinform. 2019;20(4):168. doi:10.1186/s12859-019-2684-x
- Mildrum S, Hendricks A, Stortchevoi A, et al. High-throughput miniaturized RNA-seq library preparation. J Biomol Tech. 2020;31(4):151-156. doi:10.7171/jbt.20-3104-004
- Jaeger BN, Yángüez E, Gesuita L, et al. Miniaturization of Smart-seq2 for single-cell and single-nucleus RNA sequencing. STAR Protoc. 2020;1(2):100081. doi:10.1016/j.xpro.2020.100081
- Kingan S, Wang Y, Hoffman S , et al. HiFiViral SARS-CoV-2: A mutation tolerant, fully-kitted solution for COVID-19 surveillance. https://www.sptlabtech.com. Accessed November 2022
About SPT Labtech
We Design and Manufacture Robust, Reliable and Easy-to-Use Solutions for Life Science
We enable life scientists through collaboration, deep application knowledge, and leading engineering to accelerate research and make a difference together. We offer a portfolio of products within sample management, liquid handling, and multiplexed detection that minimize assay volumes, reduce material handling costs and put the discovery tools back in the hands of the scientist.
At the Heart of What We Do
Many of our innovations have been born out of the desire to create solutions to existing customer problems; and it’s this ethos that drives SPT Labtech’s R&D efforts. Our strengths come from the trust our customers have with us to develop truly unique, automated technologies to meet their needs. We combine cutting edge science with first-rate engineering to put customers at the heart of everything we do.
A Problem-Solving State of Mind
The substantial breadth of expertise within our company enables us to be involved in the full life cycle of our products from the initial design concept, mechanical and software engineering and prototyping, to final manufacture and sale. These qualities allow us to offer the best possible technical and mechanical support to all the equipment that we supply, hence maintaining excellent client relationships.
Sponsored Content Policy: AZoLifeScience.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.