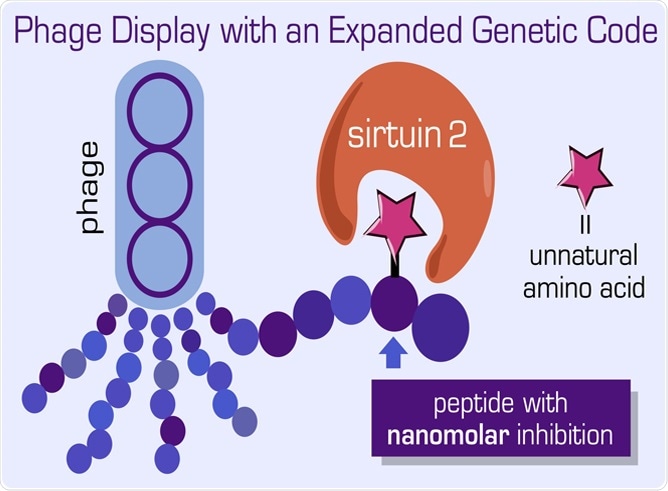
Graphic depicting peptide-protein interactions and assembly in bacteriophages. Image Credit: Dr Jeffery M. Tharp.
For many years, scientists have depended on a technique called phage display that serves as a multipurpose tool in a wide range of applications, spanning from materials science to drug discovery. The phage display technique is generally used for detecting new peptide ligands or peptides that couple to other molecules or proteins.
Headed by Dr. Wenshe R. Liu, a chemist at Texas A&M University and 2018 Presidential Impact Fellow of the same university, researchers have identified a new strategy from an old master—that is, bacteria—and effectively exploited the pathogens’ potential to create short peptides comprising noncanonical amino acids (ncAAs) that render them with unique characteristics, like targeted protein binding capabilities and enzyme degradation resistance.
The Liu research group had used an ingenious method to “trick” the system so that only those viruses that contain ncAAs-based peptides are able to reproduce.
Now, using this strategy, the researchers arranged the phage display library construction deck and effectively expanded the genetic code of bacteriophages. This method can lead to therapeutics based on novel peptides.
The researchers’ findings were published in the Nature Communications journal on March 13th, 2020.
Utilizing unnatural amino acids, we greatly expand the utility of phage display for identifying new peptide therapeutics.”
Dr Wenshe R. Liu, Chemist, Department of Chemistry, Texas A&M University
Phage display is one of the many tools used by researchers to detect novel peptides that could be used as drugs for treating various diseases, explained Dr. Jeffery M. Tharp, 2018 Texas A&M chemistry Ph.D. graduate and a postdoctoral associate at Yale University.
Dr. Tharp is also the lead author of the group’s paper. So far, this is the third paper that represents Dr. Tharp’s work at Texas A&M University. It is also one of the first papers from the Texas A&M Drug Discovery Laboratory, established by Liu and fellow Texas A&M University chemists back in 2018.
Phage display uses viruses, or phages, to ‘fish out’ specific peptides from a pool of millions of different peptide variants; however, it is very difficult to use this technique to find peptides containing ncAAs. In our paper, we developed a new method of phage display that allows for easy retrieval of potential peptide drugs containing diverse ncAAs.”
Dr Jeffery M. Tharp, Study Lead Author and PhD Graduate, Department of Chemistry, Texas A&M University
Dr. Tharp continued, “In addition, we used our new technique to identify novel peptides containing ncAAs that are very strong inhibitors of sirtuin 2—an enzyme that is involved in regulating human lifespan and is a promising drug target for the treatment of human cancers.”
The Liu research group teamed up with the Laboratory for Molecular Simulation (LMS), including LMS interim manager, Texas A&M chemistry Ph.D. candidate Andreas Ehnbom and also Texas A&M High-Performance Research Computing Associate Director Dr. Lisa M. Pérez, who carried out the molecular dynamics simulations, which allowed the researchers to interpret the selectivity involved for certain peptides.
“The beauty of this work, at least in my mind, is that it crosses multiple disciplines of chemistry—synthetic chemistry, chemical biology, and computations,” stated Ehnbom.
Dr. Tharp observed that the phage display founders received the 2018 Nobel Prize in Chemistry in recognition of the effectiveness, versatility, and relatively easy use of the technique across a wide range of disciplines.
Along with the novel resulting molecules, Dr. Tharp predicted that the technique developed by the Liu research group will equally prove handy for all phage display applications.
This technique allows ncAAs with unique structures to be incorporated into the phage peptides, which can help identify more potent peptide drugs. In addition, we can include reactive ncAAs into the phage peptides, which can potentially be used to make better materials and drug delivery systems.”
Dr Jeffery M. Tharp, Study Lead Author and PhD Graduate, Department of Chemistry, Texas A&M University
According to Dr. Tharp, the researchers will continue to apply their novel phage display method to explore other peptides containing ncAAs that inhibit the enzymes involved in human diseases and simultaneously continue to advance other techniques that would expand its usefulness.
The National Institutes of Health (Grant No. R01CA161158), the Welch Foundation (Grant No. A-1715), the Cancer Prevention and Research Institute of Texas (Grant No. RP170797), and the National Science Foundation (Grant Nos. CHE-1566601, 1664866 and 1900549) financially supported the researchers’ study.
The study titled, “An Amber Obligate Active Site-Directed Ligand Evolution Technique for Phage Display,” is available online along with related captions and figures.
Source:
Journal reference:
Tharp, J. M., et al. (2020) An amber obligate active site-directed ligand evolution technique for phage display. Nature Communications. doi.org/10.1038/s41467-020-15057-7.