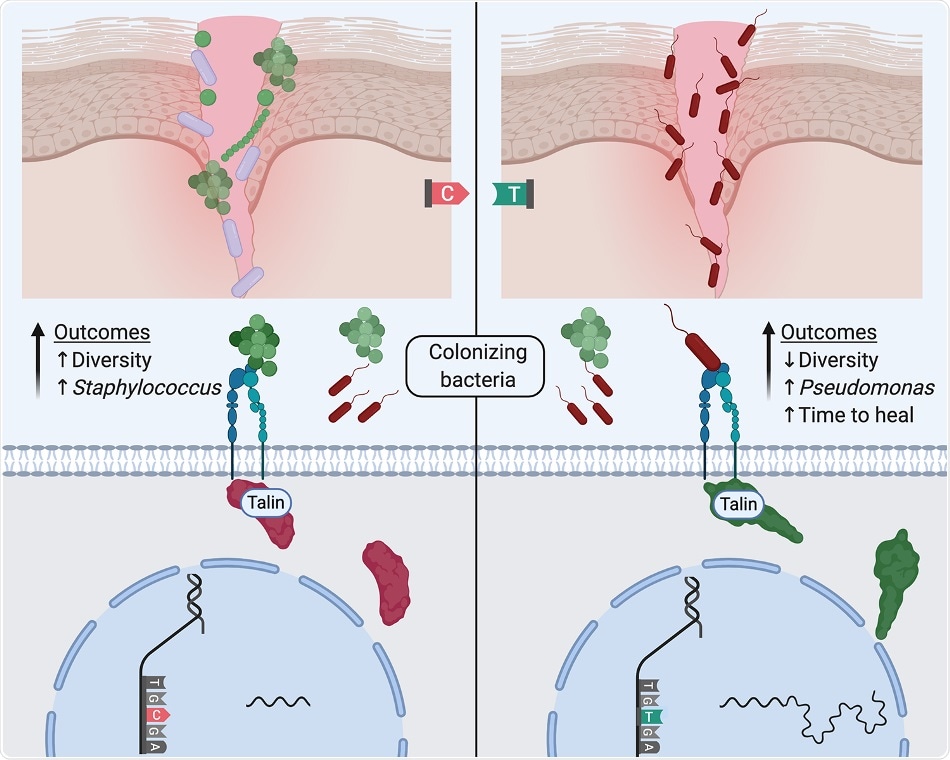
Tipton et al. provide initial insights into how patients’ genetic variation shapes cellular expression phenotypes that associate with chronic wound microbiome diversity, species abundances, and, in turn, healing. Here, a generalization of a working hypothesis emerging from the study is shown where a genetic difference influences alternative transcription of Talin2 leading to different focal adhesion dynamics that result in differential bacterial colonization of wounds. Image Credit: Texas Tech University.
If healing takes more than three weeks, physicians would call it a “chronic wound.” Such kinds of non-healing wounds impact millions of individuals and are usually related to other chronic conditions or diseases like decreased circulation, diabetes, and neuropathy.
And now, in a first-ever study, scientists have determined that genetics may play a key role in the wound healing process. The study was led by Caleb Phillips, an assistant professor at Texas Tech University and director of the Phillips Laboratory in the Department of Biological Sciences, and doctoral student Craig Tipton.
The study titled “Patient genetics is linked to chronic wound microbiome composition and healing” was published in PLOS Pathogens—an open-access, peer-reviewed medical journal—on June 18th, 2020.
According to Phillips, who also works as the Curator of Genetic Resources at the Natural Science Research Laboratory’s (NSRL) Robert J. Baker Genetic Resources Collection, the study established that specific genes are related to the abundance of common microbes and the number of bacteria present in wounds.
The collection of pathogens called a “microbiome,” can determine the process of wound healing and the time taken by that process. The study also demonstrated that the more diversity within a wound microbiome, the less time it takes the wound to heal.
A chronic wound is a serious burden. The median healing time of patients in this study was more than 200 days, but some people deal with these wounds for years. We were able to show that a person’s genetics explain differences in the species that infect their wounds.”
Caleb Phillips, Assistant Professor, Texas Tech University
Phillips continued, “The information in this study could be valuable in a clinical setting as pre-operative information to help inform preventative measures before a procedure, as some chronic wounds arise as non-healing surgical wounds, and could help inform a course of treatment for an existing infection.”
The research team included collaborators from Texas Tech University, the Texas Tech University Health Sciences Center (TTUHSC), the University of North Texas Health Science Center at Fort Worth (UNTHSC), and Lubbock’s Southwest Regional Wound Care Center (SWRWCC).
According to Phillips, the project was initiated following a discussion with Dr. Randy Wolcott, Founder of the SWRWCC, where they conversed about how particular patients develop multiple non-healing wounds across a long period. Within each wound, the microbiome was basically the same. The scientists wanted to know if this could be partly described by genetics.
This study is big. It’s the initial study to find genes and/or gene alterations that correlate with what bacterial species can be more successful in causing infections in a specific patient. If a screen of a patient's DNA prior to a surgery showed that patient is highly susceptible to a staphylococcus species, the doctor could mitigate staphylococcus complications.”
Dr Randy Wolcott, Founder, Southwest Regional Wound Care Center
Patients who visit the SWRWCC for the care of a lower-extremity infected wound agreed to participate in the research and offered samples from a cheek swab and from their wound(s). Such samples collected at the SWRWCC are generally archived in liquid nitrogen at the Wolcott Wound Care Research Collection—a collection of the NSRL’s Genetic Resources Collection particularly dedicated to wound care biology.
The study design included an exploratory group of 79 patients, from which “candidate locations” of their genome were detected. This was tracked by an experimental group of 85 patients, used to verify the links between the wound microbiome characteristics and the genomic locations.
Through microbiome sequencing techniques, the group of bacteria infecting a person’s wounds was established, and each genome of the patient was defined at a few hundred thousand specific sites, known as “single nucleotide polymorphisms,” or SNPs for short.
Later, a statistical method was employed to find out which of these genomic sites described the differences in the wound microbiome composition of an individual and was followed with many downstream analyses to interpret the outcomes.
We showed that there are identifiable locations in people's genome where, depending on their genotype, they tend to get infections by specific bacteria. The different genomic locations identified tend to be related in terms of the types of genes they are close to and may regulate.”
Caleb Phillips, Assistant Professor, Texas Tech University
Phillips continued, “A working hypothesis emerging from the research is that genetic differences influencing genes encoding the way our cells interact with the environment and each other are important for infection differences.”Tipton finished his bachelor's degree in biology at Angelo State University prior to arriving at Texas Tech University.
He stated that the project has been a major part of his thesis, which targets learning more about why an individual’s wounds are infected by a variety of microorganisms. While more work needs to be done before the research can directly benefit patients, Tipton added that the study is a promising and significant step in that direction.
According to him, “Personalized medicine is a current hot topic in modern healthcare, where the goal is to identify inherent differences within individuals that may cause them to be impacted differently by disease and finding treatments that are well-suited and tailored to the individual and may contribute to better patient outcomes.”
Tipton continued, “Our project furthers two equally-interesting avenues of research with potential translation to the clinic. In one, it is our goal to develop robust genomic predictive models that could help physicians to determine a patient's risk for chronic wound infection, particularly to specific bacteria.”
“In the second, this work helps to inform how genetic variation in patients can influence microbiome-host interactions and wound infection pathogenesis. By further studying infection pathogenesis and how these complex microbial communities interact, it may be possible to improve existing therapies or to develop new therapeutic strategies altogether,” Tipton further added.
According to Phillips, he is looking forward to pursuing his research at Texas Tech University. His laboratory is presently developing a follow-up study that he believes will gather sufficient data to build accurate predictive models. The team is also working on a study, analyzing how an individual’s location in the United States shapes variations in chronic wound microbiomes.
Caleb Phillips added, “Texas Tech provides good support for research and is continually working for growth. My research, like that of most others, has been generally enhanced by the academic freedom provided at the university. The Natural Science Research Laboratory is a premier Natural History Collection, and the samples archived at the Genetic Resources Collection have allowed me to design studies such as this one that would otherwise not have been possible.”
“The hard work and creativity of doctoral student Craig Tipton were essential to the success of this project, as was a collaboration with the laboratories of Nicole Phillips at UNTHSC and Kendra Rumbaugh at TTUHSC, Professor Todd Little in the Texas Tech College of Education, the SWRWCC and the NSRL,” Phillips concluded.
Source:
Journal reference:
Tipton, C. D., et al. (2020) Patient genetics is linked to chronic wound microbiome composition and healing. PLOS Pathogens. doi.org/10.1371/journal.ppat.1008511.