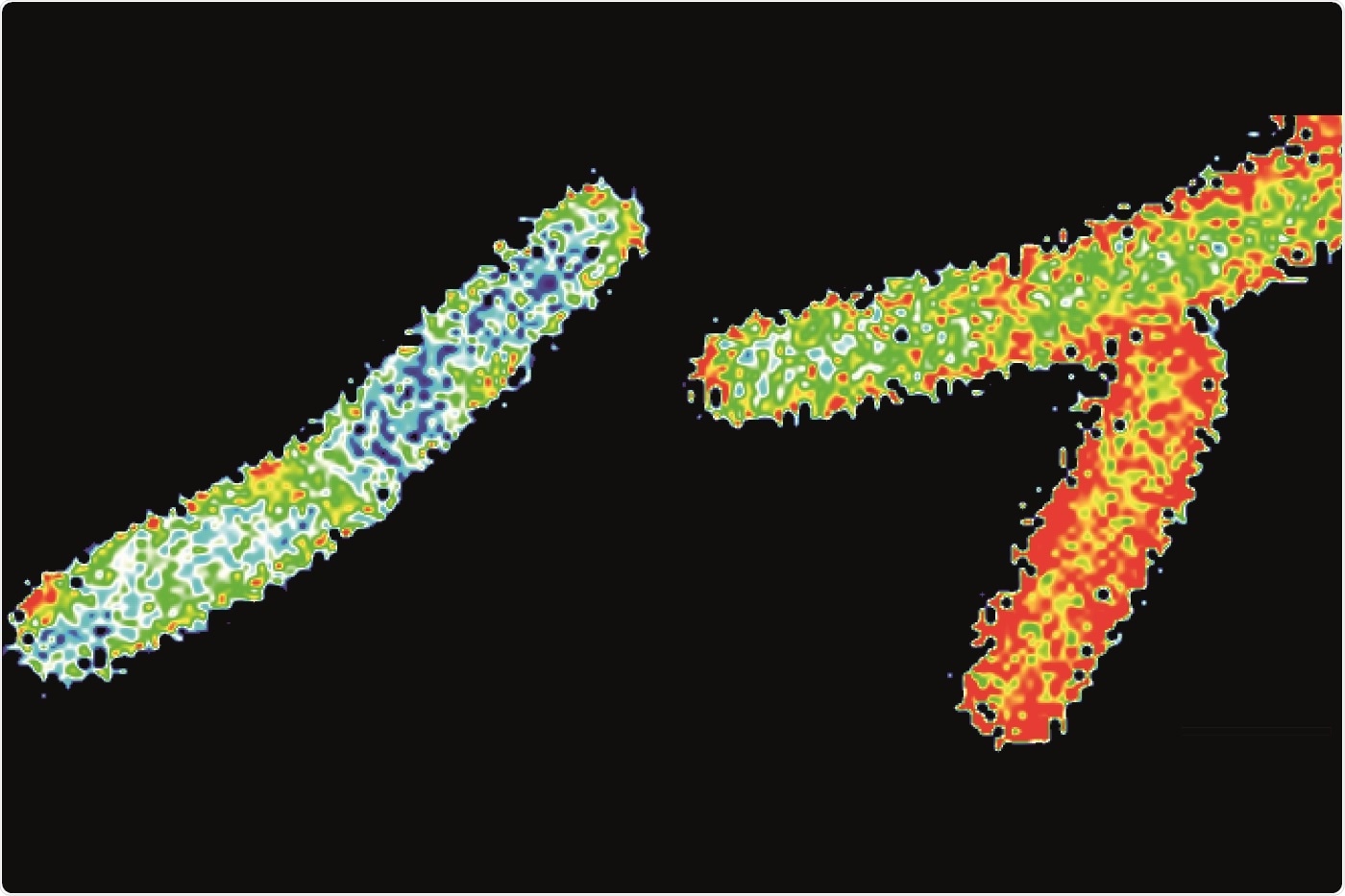
The membrane of Bacillus subtilis cells was marked with the dye Laurdan and analyzed by fluorescence microscopy. The staining indicates the fluidity of the membrane: The membranes of the blue-stained cells are considerably more rigid than the very fluid membranes of the reference cells (in red). Image Credit: © Professor Marc Bramkamp.
To apply those characteristics, different kinds of proteins are active in cells, for example, the supposed flotillins. Such proteins are found in cells ranging from humans to bacteria.
To date, researchers have believed that such flotillins primarily help in the development of other functional protein complexes and restrict highly ordered regions of the cell membrane.
Now, an international research team, including scientists from Kiel University, has identified indications of a potential function of the flotillins.
Along with collaborators from the University of Groningen in the Netherlands and the University of Bordeaux in France, among others, researchers from Kiel University have successfully demonstrated that flotillin proteins evidently have a direct impact on the cell membrane structure and can render it more fluid under certain situations.
The researchers recently published their results in eLife, a leading scientific journal.
Flotillins may work differently than previously assumed
Every living cell should be surrounded by a separating obstacle that protects them from their setting and yet should also be permeable for numerous molecular substances. Different proteins are required to form the cell membrane and to provide them with their functions.
To date, scientists have believed that the supposed flotillin proteins play a role in the formation of the required functional protein complexes—for instance, by delimiting the specific regions of the membrane. The study, which was recently presented by Professor Marc Bramkamp’s Microbial Biochemistry and Cell Biology team at the Institute of General Microbiology in Kiel University, challenges this view.
Together with a group of international colleagues, we have found evidence that the flotillin proteins have a completely different function. Apparently, they regulate the fluidity of bacterial membranes, making them more fluid to a certain extent and thus, changing their properties.”
Marc Bramkamp, Professor, Institute of General Microbiology, Kiel University
Such an assumption may also explain the impact that flotillins have on the development of cell wall: For the cell wall, building blocks are created in the cells’ interior and should later be “flipped” outwards, which is much easier to perform in a more fluid membrane.
Additionally, the protein machinery that produces the cell wall moves dynamically via the cell membrane and this locomotion is considerably decreased without the presence of flotillins. As a result, the cells cannot synthesize the cell wall properly.
Without flotilline, no stable form
In association with international collaborators, the research group from Kiel University developed a novel hypothesis based on experiments performed with the rod-shaped bacterium known as Bacillus subtilis. Such experiments demonstrated that the rapidly-growing cells are incapable of developing their characteristic shape without the presence of flotillins.
But if the scientists introduced a chemical substance to fluidize the membranes, the pathogens could preserve their shape even in the absence of flotillin proteins.
We therefore assume that they take over a physical role in the bacterial membrane. The flotillins seem to have an effect on the physical structure of the membrane, conferring the correct fluidity so other membrane-bound processes can function properly.”
Abigail Savietto, PhD Student, Kiel University
Savietto is part of Bramkamp’s group.
In upcoming studies, the scientists are hoping to establish the exact molecular mechanism between the membrane fluidity and flotillin proteins. One method might be the analysis of the membrane’s phospholipid composition.
Such lipids play a role in the formation of several different biomembranes. Flotillin proteins could adhere to certain phospholipids that decrease fluidity and thus boost the overall fluidity of the cell membrane.
Therefore, the new theory proposed by Kiel University researchers also holds promising implications for other applications: in the days to come, the physical properties of bacterial cell membranes could be particularly influenced by interrupting the role of flotillins.
My research group has been working on the function of flotillins for many years and we know that cells with altered membrane fluidity are much more sensitive to conventional antibiotics. It might be possible to use this mechanism, for example to specifically alter the membrane of bacterial cells in such a way that they can be killed more easily with antibiotics.”
Marc Bramkamp, Professor, Institute of General Microbiology, Kiel University
Source:
Journal reference:
Zielińska, A., et al. (2020 Flotillin-mediated membrane fluidity controls peptidoglycan synthesis and MreB movement. eLife. doi.org/10.7554/eLife.57179.