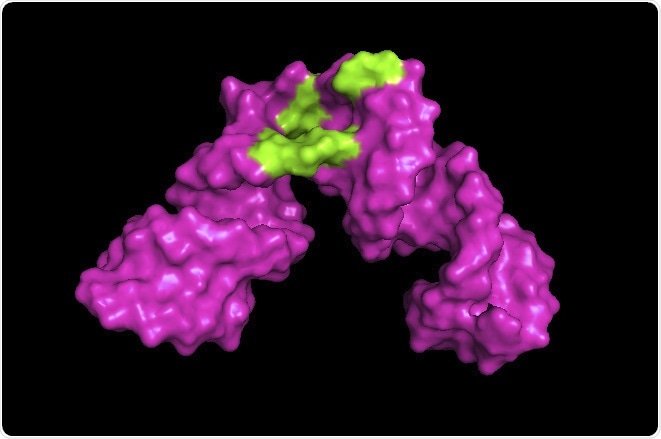
University of Maryland researchers developed a method to expand the scope of nuclear magnetic resonance (NMR) spectroscopy. In the example above, the researchers were able to create a 3D image revealing the site on a piece of hepatitis RNA where small molecules such as a drug could bind (shown in green). Image Credit: Kwaku Dayie/University of Maryland.
The novel approach extends the scope of nuclear magnetic resonance (NMR) spectroscopy and will allow scientists to interpret the structure and shape of RNA molecules and learn their mode of interaction with other molecules.
The technology offers insights that may lead to targeted RNA therapeutic treatments for arious diseases. A research article on this latest work was published in the Science Advances journal on October 7th, 2020.
The field of nuclear magnetic resonance spectroscopy has been stuck looking at things that are small, say 35 RNA building blocks or nucleotides. But most of the interesting things that are biologically and medically relevant are much bigger, 100 nucleotides or more. So, being able to break down the log jam and look at things that are big is very exciting. It will allow us to peek into these molecules and see what is going on in a way we haven’t been able to do before.”
Kwaku Dayie, Study Senior Author and Professor, Department of Chemistry and Biochemistry, University of Maryland
In nuclear magnetic resonance (NMR) spectroscopy, investigators direct radio waves at a specific molecule, stimulating the atoms and “lighting up” that molecule. By quantifying the variations in the magnetic field around the excited atoms—the nuclear magnetic resonance—researchers can rebuild characteristics, like the structure, shape, and movement of the molecule. This produces data that can be subsequently used to generate pictures, almost like MRI images observed in medicine.
Generally, NMR signals emitted from several atoms in a biological molecule, like RNA, tend to overlap with one another, which makes analysis highly complicated. But in the 1970s, investigators learned to biochemically engineer RNA molecules to function better with NMR by substituting the hydrogen atoms with magnetically active fluorine atoms.
In comparatively small RNA molecules containing 35 or fewer nucleotides, the fluorine atoms illuminate instantly when struck with radio waves and continue to remain excited to enable high-resolution analysis.
However, as RNA molecules become larger, the fluorine atoms illuminate only for a short time and then rapidly lose their signal. This has inhibited high-resolution 3D analysis of larger molecules of RNA.
Earlier studies performed by other scientists had demonstrated that fluorine continued to create a powerful signal when it was located next to a carbon atom consisting of seven neutrons and six protons (C-13).
Therefore, Dayie and his group devised a relatively simple technique to modify the naturally occurring C-12 in RNA (which contains six neutrons and six protons) to C-13 and install a fluorine atom (F-19) directly adjacent to it.
Dayie and his group initially showed that their technique can generate images and data that are equivalent to present-day techniques by applying it to RNA pieces from HIV containing 30 nucleotides, which had been already imaged. The team subsequently applied their technique to parts of Hepatitis B RNA comprising 61 nucleotides—almost twice the size of earlier NMR spectroscopy possible for RNA.
The team’s method allowed the investigators to detect the locations on the hepatitis B RNA in which small molecules attach and interact with the RNA. That might prove handy for interpreting the impact of promising therapeutic drugs. The subsequent step for the scientists is to examine even larger molecules of RNA.
This work allows us to expand what can be brought into focus. Our calculations tell us that, in theory, we can look at really big things, like a part of the ribosome, which is the molecular machine that synthesizes proteins inside cells.”
Kwaku Dayie, Study Senior Author and Professor, Department of Chemistry and Biochemistry, University of Maryland
By interpreting the structure and shape of a molecule, investigators can gain a deeper understanding of its function and its interaction with the environment. Moreover, this technique will allow scientists to observe the 3D structure as it modifies, since RNA molecules specifically alter their shape frequently. This understanding is essential for developing therapeutics that closely target disease-specific molecules without causing an impact on the function of healthy cells.
The hope is that if researchers know the nooks and crannies in a molecule that is dysfunctional, then they can design drugs that fill the nooks and crannies to take it out of commission.”
Kwaku Dayie, Study Senior Author and Professor, Department of Chemistry and Biochemistry, University of Maryland
“And if we can follow these molecules as they change shape and structure, then their response to potential drugs will be a little bit more predictable, and designing drugs that are effective can be more efficient,” Dayie concluded.
Source:
Journal reference:
Lhee, S., et al. (2020) Spatial localization of charged molecules by salt ions in oil-confined water microdroplets. Science Advances. doi.org/10.1126/sciadv.aba0181.