An international research alliance has visualized the RNA folding structures of the SARS-CoV-2 genome, for the first time. The SARS-CoV-2 virus uses this genome to regulate the infection process.
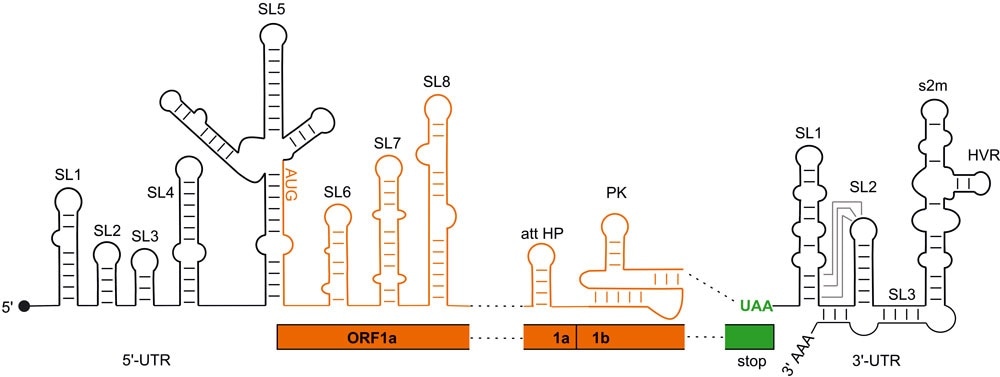
Regulatory RNA elements of the SARS-CoV2 genome. Black: regions not coding for proteins (UTR); orange: coding regions (ORF). Image Credit: Goethe University Frankfurt.
Since such structures are extremely analogous among numerous beta corona viruses, the researchers laid the basis for the targeted development of new drugs for COVID-19 treatment and also for upcoming occurrences of infection with novel coronaviruses that may evolve in the days to come.
Within the SARS-CoV-2 virus, the genetic code is precisely 29,902 characters long and strung through an extended RNA molecule. It includes data for producing 27 proteins. This is very little when compared to the potential 40,000 kinds of protein that can be produced by a human cell.
But viruses employ the metabolic processes of their host cells to grow in numbers. Viruses that can accurately regular the synthesis of their individual proteins are vital to this strategy.
SARS-CoV-2 utilizes the spatial folding of its RNA hereditary molecule as a control element for producing proteins—mainly in regions that do not code for the viral proteins, Single strands of RNA adopt structures with RNA double strand loops and sections. But the only models of these folding have been built on indirect experimental evidence and computer analyses, until now.
For the first time, an international research team, headed by chemists and biochemists from Goethe University and TU Darmstadt, have now experimentally tested the models. The study also involved scientists from the Israeli Weizmann Institute of Science, the Swedish Karolinska Institute, and the Catholic University of Valencia.
The team successfully defined the structure of a total of 15 of these regulatory elements. They achieved this by using nuclear magnetic resonance (NMR) spectroscopy, wherein the atoms of the RNA are subjected to a powerful magnetic field, and thus demonstrated something about their spatial arrangement.
The researchers compared the latest findings from this technique with the findings observed from a chemical process (dimethyl sulfate footprint), which enables the regions of RNA single strands to be differentiated from the regions of RNA double strands.
Our findings have laid a broad foundation for future understanding of how exactly SARS-CoV-2 controls the infection process. Scientifically, this was a huge, very labor-intensive effort which we were only able to accomplish because of the extraordinary commitment of the teams here in Frankfurt and Darmstadt together with our partners in the COVID-19-NMR consortium.”
Harald Schwalbe, Professor and Coordinator of the Consortium, Center for Biomolecular Magnetic Resonance, Goethe University Frankfurt
Schwalbe continued, “ But the work goes on: together with our partners, we are currently investigating which viral proteins and which proteins of the human host cells interact with the folded regulatory regions of the RNA, and whether this may result in therapeutic approaches.”
Globally, more than 40 working teams with 200 researchers are performing studies within the COVID-19-NMR consortium, including 45 doctoral students and postdoctoral students in Frankfurt working in two shifts daily, seven days of the week since the end of March 2020.
Schwalbe is satisfied that the possibility for the discovery goes beyond novel therapeutic alternatives for SARS-CoV-2 infections.
The control regions of viral RNA whose structure we examined are, for example, almost identical for SARS-CoV and also very similar for other beta-coronaviruses. For this reason, we hope that we can contribute to being better prepared for future ‘SARS-CoV3’ viruses.”
Harald Schwalbe, Professor and Coordinator of the Consortium, Center for Biomolecular Magnetic Resonance, Goethe University Frankfurt
Source:
Journal reference:
Wacker, A., et al. (2020) Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy. Nucleic Acid Research. doi.org/10.1093/nar/gkaa1013.