A genetically encoded “live-cell” probe developed by scientists from Tokyo Tech pinpointed phosphorylated Ser2 in the enzyme RNA polymerase II, significantly determining sites of the elongation phase of active transcription in live cells. This probe can be utilized to image transcription elongation in real-time in living animals, enhancing further breakthrough gene regulation studies.
Live imaging of transcription sites using an elongation RNA polymerase II-specific probe. Image Credit: Uchino et al., 2021| Journal of Cell Biology.
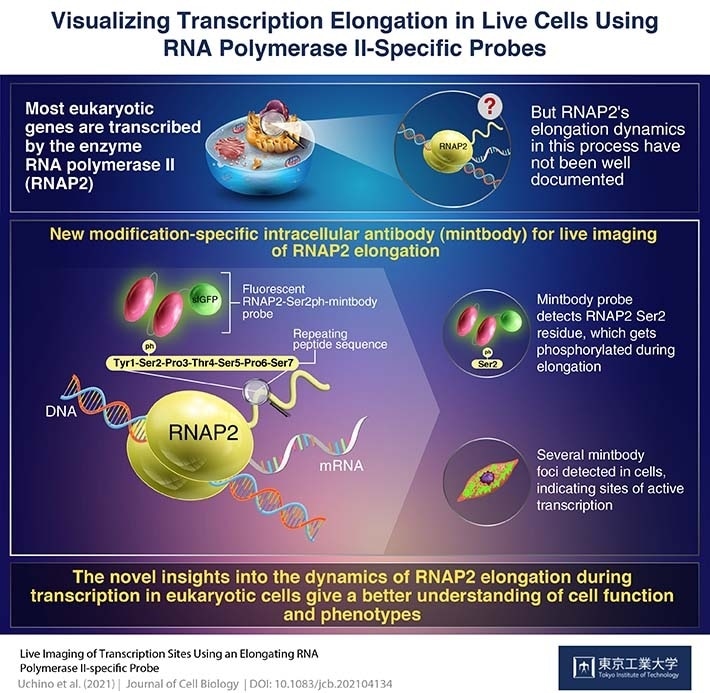
The nuclei of all eukaryotic cells contain an enzyme called RNA polymerase II (RNAP2) that is responsible for transcribing mostly all the genes that offer the various characteristics of humans.
Hence, it is important to get a better understanding of the complete eukaryotic genome regulation and the functions of different cells—RNAP2 behavior in live cells was examined utilizing fluorescent protein-tagged RNAP2 molecules. Although this approach increased the understanding of the protein’s behavior, a cell generally has more than 100,000 RNAP2 molecules and thousands of active transcription sites or “factories”.
It is a tedious process to track single RNAP2 foci when the whole-cell lights up like a lightbulb due to fluorescence!
However, lately, a process of live-cell visualization engaging the use of fluorescently tagged antigen-binding fragments achieved localization of, particularly modified protein forms.
Taking advantage of this approach, a team of scientists from Tokyo Institute of Technology and Kyushu University, headed by Professor Hiroshi Kimura, created a genetically encoded “live-cell” probe to track a modified version of RNAP2 that is seen in cells at the time of transcription. The study was published in the Journal of Cell Biology.
Professor Kimura details their rationale very plainly.
In humans, most genes are transcribed by RNAP2, which is a 12-unit complex structure. The largest subunit has a long tail consisting of 52 repeats of seven amino acids. During the transcription elongation phase, one of these seven amino acids, Ser2, is phosphorylated. We attempted to detect this Ser2-phosphorylated, active RNAP2 in living cells.”
Hiroshi Kimura, Professor, Tokyo Institute of Technology
A genetically encoded probe is convenient while introducing them into cells when compared to protein probes, as the cells go on producing it after the genetic material encoding the probe integrates with the host DNA. Professor Kimura’s group created a genetically encoded modification-specific intracellular antibody (indicated as “mintbody”) probe, named RNAP2-Ser2ph-mintbody, to identify RNAP2 Ser2 phosphorylation.
Upon investigating the cells with high-resolution microscopy, the researchers discovered the presence of the RNAP2-Ser2ph-mintbody in regions containing active transcription.
Numerous mintbody foci were found, almost like transcription ‘factories.’ This could indicate that the RNAP2-Ser2ph-mintbody can recognize active transcription sites.”
Hiroshi Kimura, Professor, Tokyo Institute of Technology
The scientists noted that the RNAP2-Ser2ph-mintbody reduced during cellular mitosis and in the presence of RNAP2 inhibitors. They also found that the RNAP2-Ser2ph-mintbody turned up frequently when associated with other proteins involved in the elongation phase of transcription, compared to the proteins engaged in the initiation and termination phases.
The RNAP2-Serph2 foci in the cells were comparatively more mobile than DNA replication domains, indicating that loci, where transcription elongation occurs, are detached and more mobile than limited chromatin domains.
What does this indicate for genetic research’s future?
Understanding the regulation of RNAP2 can help us understand genomes, cell functioning, and phenotypes.”
Hiroshi Kimura, Professor, Tokyo Institute of Technology
Undoubtedly, transcription is the initial step to protein synthesis, and localizing sites of active transcription provides a target to investigate the mechanism. Thorough knowledge of how DNA is read by RNAP2 could offer important clues about the outcome of protein synthesis.
The findings of this research can encourage future research in genetics and gene regulation and expand the understanding of the human genome.
Source:
Journal reference:
Uchino, S., et al. (2021) Live imaging of transcription sites using an elongating RNA polymerase II–specific probe. Journal of Cell Biology. doi.org/10.1083/jcb.202104134.