Reviewed by Danielle Ellis, B.Sc.Jan 8 2024
Based on their amino acid sequences, proteins fold into distinct three-dimensional structures that determine their function. While de novo protein design has advanced significantly, complex all-αproteins, in which α-helices are not stacked in a parallel fashion within three-dimensional structures, have not been as easily designed.
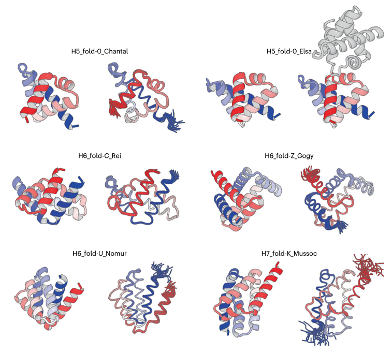 De novo designed all-α proteins with complicated shapes: H5_fold_0_Chantal, H5_fold-0_Elsa, H6_fold-C_Rei, H6_fold-Z_Gogy, H6_fold-U_Nomur, and H7_fold-K_Mussoc are presented. For each de novo-designed protein, the computational model is shown on the left, and the solved experimental structure is on the right. Image Credit: Osaka University
De novo designed all-α proteins with complicated shapes: H5_fold_0_Chantal, H5_fold-0_Elsa, H6_fold-C_Rei, H6_fold-Z_Gogy, H6_fold-U_Nomur, and H7_fold-K_Mussoc are presented. For each de novo-designed protein, the computational model is shown on the left, and the solved experimental structure is on the right. Image Credit: Osaka University
Prof. Nobuyasu Koga of the Exploratory Research Center on Life and Living Systems (ExCELLS) at the National Institutes of Natural Sciences (NINS) says, “Artificially designed proteins mostly show simple structures, but Nature presents us with complicated ‘designs.’” This discrepancy motivated the group to look for a way to create such intricate all-α proteins.
Image Credit: StudioMolekuul/Shutterstock.com
Using structures that were deposited in the Protein Data Bank (PDB), the team first identified 18 common helix-loop-helix motifs. They then showed that by combining these discovered characteristic motifs with canonical α-helices, a wide range of all-α protein tertiary structures, from basic to complex, may be computationally produced.
It is surprising that such a diverse set of all-α protein structures can be generated simply by combining typical or canonical components of naturally occurring proteins.”
Dr Koya Sakuma, Former PhD Student, SOKENDAI (The Graduate University for Advanced Studies).
From the computationally produced structures, the team picked five distinct all-α proteins, each having five or six α-helices and unique in their intricate spatial configurations. Subsequently, they designed amino acid sequences from scratch such that they could fold into the five all-α protein structures that were chosen. The experimental findings were astounding.
The structures that emerged from the structure determination process exhibit the complicated shapes, which perfectly matched computationally designed structures.”
Dr Naohiro Kobayashi, Senior Research Fellow, RIKEN
It is now feasible to produce complexly shaped all-α proteins using this newly established technique. Protein three-dimensional architectures are intrinsic to their functions. Creating complexly shaped proteins enhances the likelihood of creating functional places within them. New functional proteins will be made possible by this ground-breaking discovery, which will advance the life sciences and healthcare.
Source:
Journal reference:
Sakuma, K., et.al., (2024). Design of complicated all-α protein structures. Nature Structural & Molecular Biology. doi.org/10.1038/s41594-023-01147-9.