Reviewed by Danielle Ellis, B.Sc.Nov 27 2024
To better understand the highly varied characteristics that enable malaria parasites to adhere to red blood cells and elude the immune system, researchers have developed a new tool.
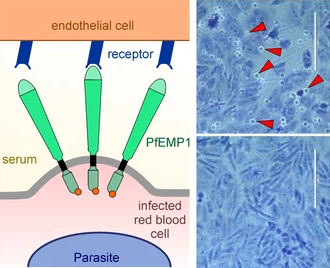 Left, schematic showing section of a red blood cell infected with a malaria parasite binding to the endothelium of the human host via PfEMP1. Right, microscopy image of a binding assay: infected red blood cells binding to cells expressing endothelial receptor (top) or control cells not expressing the receptor (bottom); red arrows, examples of bound infected red blood cells. Image credit: Tobias Spielmann
Left, schematic showing section of a red blood cell infected with a malaria parasite binding to the endothelium of the human host via PfEMP1. Right, microscopy image of a binding assay: infected red blood cells binding to cells expressing endothelial receptor (top) or control cells not expressing the receptor (bottom); red arrows, examples of bound infected red blood cells. Image credit: Tobias Spielmann

 *Important notice: eLife publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: eLife publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
The study, released in eLife, presents a significant method for creating Plasmodium falciparum parasite lines that express specific variants of a sticky adhesin molecule. They also highlight that the research offers strong evidence for an innovative and robust platform to investigate the mechanisms through which malaria causes disease.
The human malaria parasite's cytoadhesion the capacity to adhere to red blood cells allows it to evade immune system clearance and result in the accumulation of infected red blood cells in vital organs, which can have fatal outcomes.
Members of the protein family known as “P. falciparum erythrocyte membrane protein 1,” or PfEMP1, aid in cytoadhesion. Each parasite can flip between multiple forms of PfEMP1 to evade immune detection, but it only produces one variety of PfEMP1 at a time, controlled by its genes.
A key problem in studying PfEMP1 is that when you grow malaria in the lab, it results in a population of parasites with diverse PfEMP1 proteins with different properties. Researchers have tried to select for parasite strains producing certain PfEMP1 proteins or use antibodies to study them, but this has proved challenging.”
Jakob Cronshagen, Study Lead Author and PhD Student, Bernhard Nocht Institute for Tropical Medicine
Jakob Cronshagen was a Ph.D. Student shared between the Bruchhaus and Spielmann labs at the Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany.
To solve this, Cronshagen and associates employed a technique known as selection-linked integration (SLI), which involves selectively enriching parasites with a particular kind of protein using an antibiotic. This allowed the team to better examine aspects of PfEMP1 biology by producing parasites that primarily produce a single PfEMP1 protein.
For instance, prior research has shown that mutations of several proteins involved in PfEMP1 trafficking prevent its transport at various stages as it travels to the surface of red blood cells. By obtaining parasites that all expressed a tagged form of PfEMP1, the authors were able to track its movements using their novel SLI method.
The PfEMP1 “proxiome,” or the collection of proteins that work most closely with PfEMP1 during malaria infection, was also studied by the authors using their novel approach. In addition to validating existing relationships, they discovered two new proteins that are involved in the cytoadhesion process.
Allowing the parasite to transition between PfEMP1 variations, as it would naturally do during developing malaria infection, is another crucial prerequisite for researching these harmful PfEMP1 proteins.
The parasites changed from a homogeneous population that mostly produced the same PfEMP1 to a diversified population that produced a variety of PfEMP1 variations, according to the team's test, which involved eliminating the antibiotic that was used to select for the particular PfEMP1.
The way in which PfEMP1 functions and how it is neutralized by human antibodies are increasingly being uncovered on a structural level and are essential for understanding malaria pathology and immune responses in patients. The straightforward capacity to generate cell-adherent parasite lines uniformly expressing a single PfEMP1 of interest will open up new approaches to block these pathogenicity proteins as a new therapeutic strategy in malaria.”
Tobias Spielmann, Research Group Leader and Study Senior Author, Bernhard Nocht Institute for Tropical Medicine

 *Important notice: eLife publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: eLife publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Source:
Journal reference:
- Preliminary scientific report.
Cronshagen, J., et al. (2024) A system for functional studies of the major virulence factor of malaria parasites. eLife. doi.org/10.7554/elife.103542.1.