Microalgae are photosynthetic organisms that live in water systems and convert atmospheric CO2 to organic molecules, including lipids, carbohydrates, and other bioactive compounds.
With such biological properties, microalgae are a sustainable and renewable source of food ingredients, providing animal-free omega-3, an alternative source of protein, and natural pigments for the food and beverage division.
The great promise shown by microalgae to meet dietary needs has garnered much attention from the food industry.
According to the European Food Safety Authority (EFSA), the demand for natural pigment is increasing among consumers who prefer natural products to potentially toxic synthetic food additives.1
However, natural pigments may be light-sensitive and can experience color change following exposure to different environmental conditions (such as pH and temperature). They are also usually available in a limited color range.
These factors motivate the research and development around using new natural pigments as food dyes.
High throughput automation is required to screen many microalgal strains in minimal time to facilitate the discovery of microalgal pigments with valuable properties for the food industry.
Fermentalg® laboratories have utilized high throughput automation and leveraged Galdieria sulphuraria metabolism to develop a natural blue pigment rich in C-phycocyanin. This is extracted and commercialized as BLUE ORIGINS®.2
BLUE ORIGINS® is a natural pigment that is stable at low pH and high temperatures relative to other microalgae-derived dyes. This means it is suitable for various applications in the food and beverage industry, such as ready-to-eat cereals, candies, and beverages.
C-phycocyanin is the only blue pigment in the FDA food coloring agent-exempt product list. This means it is appropriate for use in manufacturing natural products.3
Fermentalg® scientists have created innovative methods for screening and selecting microalgal strains with the QPix 420 microbial colony picker (Molecular Devices®).
Traditionally, colony picking is a manual process that involves sterile pipette tips or inoculation loops, making it both time-consuming and labor-intensive.
The QPix 420 system enables high throughput colony picking, and it can pick more than 3,000 colonies within an hour with a picking efficiency higher than 98%.
A range of fluorescent filter sets enhances system flexibility and walk-up operations, allowing the phenotypic selection of unique colonies based on morphological features and fluorescent protein expression levels or fluorescent pigments.
Benefits of Fluorescence on QPix® 420
- Quantitative fluorescent screening allows efficient and objective selection of unique clones.
- It maintains subsequent clonal integrity during bio-production
- A variety of fluorescent filter sets improves experimental flexibility
- Easy-to-use software means that the right colony is picked based on the defined experimental parameters
Materials and Methods
In the search for a strain of Galdieria sulphuraria producing high levels of C-phycocyanin, Fermentalg® has obtained and preserved a set of variants with different C-phycocyanin contents. The strains were plated on Gross-agar medium4 (pH 3) and incubated for two weeks at 37 °C with 60% humidity in the dark.
All plates were imaged and analyzed with the QPix 420 system, allowing fast walk-up operations. Compatibility with different microorganisms is ensured using an appropriate pin type; in this case, Christmas tree-shaped pins were utilized to successfully pick and inoculate G. sulphuraria colonies.
Sterility was maintained using three wash baths to sanitize pins (as shown in Figure 1).
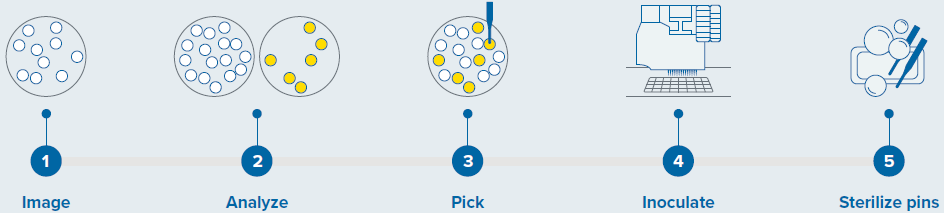
Figure 1. Colony screening workflow with QPix 420 system. Image Credit: Molecular Devices UK Ltd
Results
A Fluorescent Approach to Screening G. Sulphuraria Colonies
A fluorescence-based technique was employed to select colonies of the microalgae Galdieria sulphuraria, rich in C-phycocyanin, a water-soluble protein in blue-green algae responsible for their characteristic bright blue color.
When subjected to a red light, C-phycocyanin emits fluorescence with a peak at 642 nm.5
Colonies of G. sulphuraria that were two weeks old were imaged and screened using transmitted light and fluorescence on the QPix 420 system. The system allows the objective identification and selection of colonies that produce the fluorescent pigment of interest (C-phycocyanin).
A test image of the plate under transmitted light and the selected fluorescence is attained with the QPix Fusion Software (as displayed in Figure 2A and B, respectively). These test images can then be altered by specifying the acquisition settings, including exposure time.
The detected colonies can be visualized by a green overlay produced by the software algorithm in correspondence with every detected feature.
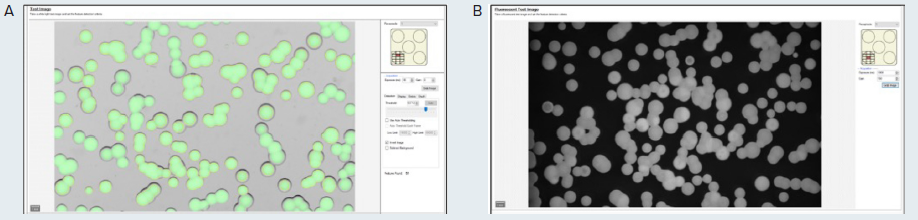
Figure 2. (A) Test image in transmitted light of the microalgae G. sulphuraria. QPix Fusion Software™ shows detected features creating a green overlay on each colony. (B) Test image of the same sample was acquired in fluorescence channel (Ex/Em filter: 628/692nm) to detect colonies expressing C-phycocyanin. Image Credit: Molecular Devices UK Ltd
Following the capture of these images, the QPix Fusion Software analyzes the images based on the feature properties, identifying the location of individual colonies on the culture plate (Petri plate, Omnitray, or QTray).
The Fusion software flexibility allows users to define the desired colonies based on morphology, such as axis ratio, colony size, diameter, compactness, and minimum proximity.
Subsequently, selected colonies of G. sulphuraria were either included (as presented in Figure 3, yellow) or excluded (as presented in Figure 3, orange) based on the selection criteria.
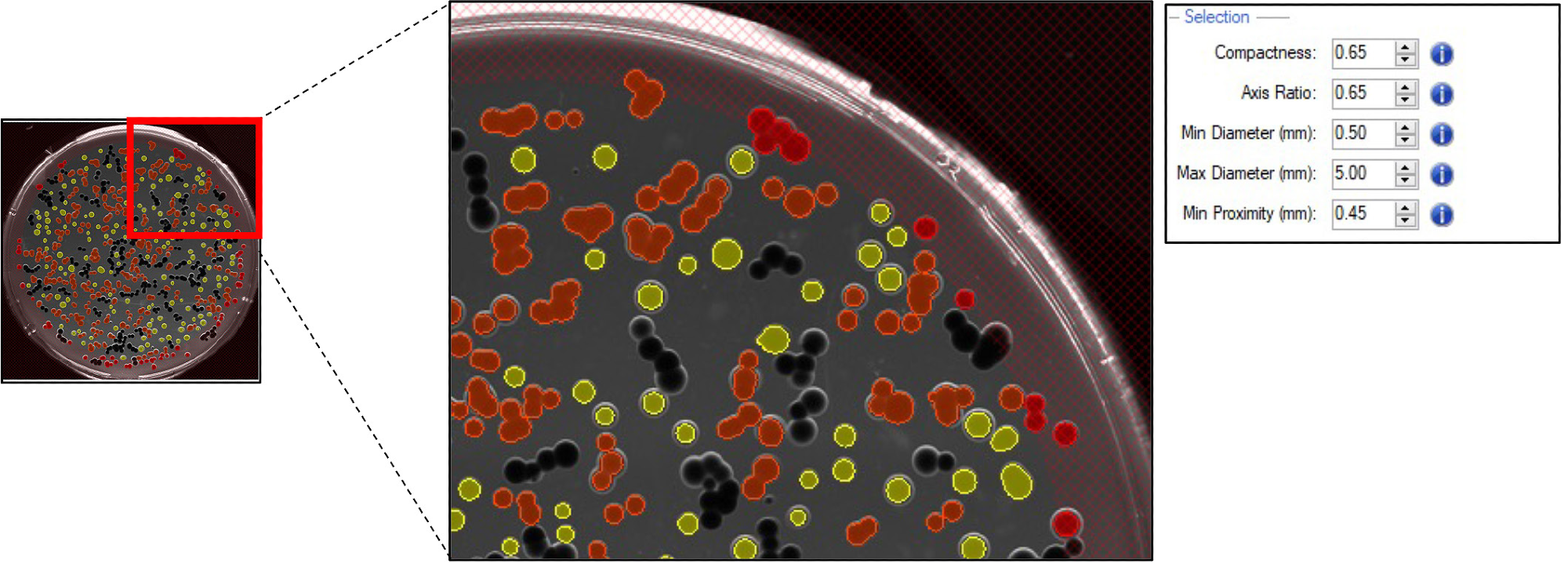
Figure 3. Pickable colonies are displayed in yellow. Orange: colonies excluded according to selection criteria. Top right panel: selection criteria defined for accurate colony picking. Image Credit: Molecular Devices UK Ltd
The quantitative fluorescence intensity threshold may be tuned to specify the desired protein expression level. To select cells with the greatest concentrations of C-phycocyanin, the range of fluorescence needs to be adjusted.
For this experiment, colonies that produce C-phycocyanin with a Mean Fluorescence Intensity (MFI) value between 36,000 and 43,000 were selected (as shown in Figure 4A and B, respectively). Once selected based on the criteria defined by the user, colonies were accurately picked using the QPix 420 system.
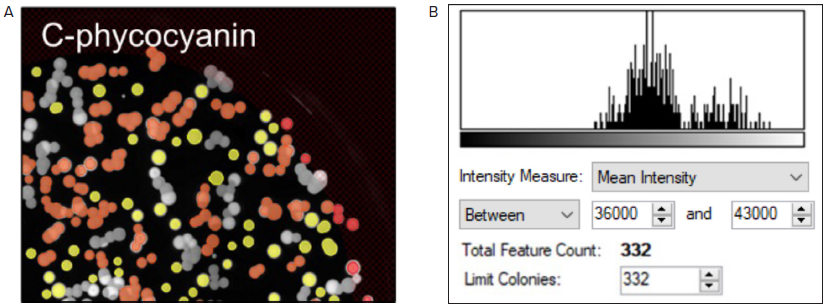
Figure 4. Fluorescent screening of G. sulphuraria colonies producing C-phycocyanin. (A) Representative fluorescent image of G.sulphuraria colonies expressing different levels of C-phycocyanin. The Red filter pair (Ex/Em: 628/692) was used. (B) Fluorescence histogram displayed as Mean Intensity. Image Credit: Molecular Devices UK Ltd
Screening and Detecting High-Expressing C-Phycocyanin Colonies
The QPix 420 system was employed to analyze various G. sulphuraria strains that produce different concentrations of C-phycocyanin to test the ability of the equipment to distinguish and select the high-expressing C-phycocyanin colonies, which appear as dark green (as shown in Figure 5A).
As expected, the colonies with the lowest concentration of C-phycocyanin appeared white on agar plates, and the colonies that expressed average pigment levels appeared light green.
The different strains were imaged via transmitted light with the QPix 420 and subsequently screened based on C-phycocyanin fluorescent emission (Figure 5B-C).
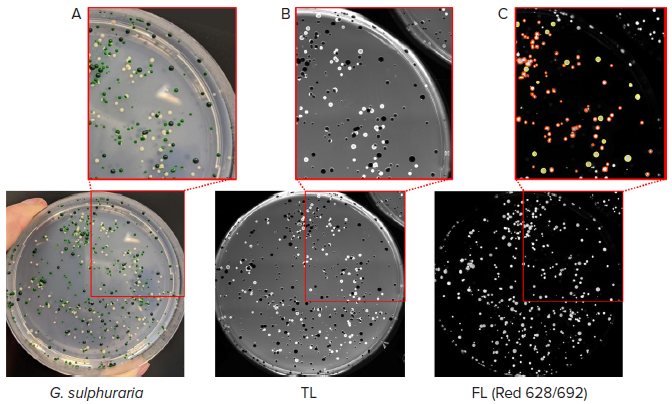
Figure 5. Selection of high-expressing C-phycocyanin colonies. (A) Colonies of G. sulphuraria with different pigmentation were analyzed. (B) Transmitted light (TL) image of the same plate of colonies detected with QPix 420 camera. (C) Fluorescent image (Red filter pair, Ex/Em: 628/692nm) of G. sulphuraria colonies. Pickable colonies are displayed in yellow. Orange: colonies excluded according to selection criteria. Image Credit: Molecular Devices UK Ltd
A fluorescent signal for the light and dark green-colored colonies was detected. Dark green colonies were selected and picked using the previously tested Mean Fluorescence Intensity values within the range of 36,000 and 43,000.
The disappearance of the white colonies in fluorescent imaging confirmed the absence of C-phycocyanin in this microalgae population.
Conclusion
The food industry’s interest in microalgae is increasing due to the growing demand for natural and sustainable ingredients.
Fermentalg® leverages microalgae metabolism to produce and screen multiple G. sulphuraria strains that are stable at a low pH and high temperature and can act as a natural blue pigment for food ingredients (BLUE ORIGINS®).
Automated colony screening of G. sulphuraria colonies was carried out to accelerate this product to the industry. High throughput colony picking delivers considerable advantages compared to conventional screening methods, such as faster identification of desired clones to streamline the following downstream analysis.
The QPix 420 fluorescence abilities were utilized to select clones that produce the greatest concentrations of C-phycocyanin, increasing the speed of the scale-up of microalgae-based natural pigment production. High throughput automation has great potential for the future development of microalgal products.
Acknowledgments
Produced from materials originally authored by Carola Mancini from Molecular Devices, Audrey S. Commault, Rodrigo Rangel, and Julien Demol from Fermentalg.
References and Further Reading
- Carocho M, et al. Adding Molecules to Food, Pros and Cons: A Review on Synthetic and Natural Food Additives. Compr Rev Food Sci Food Saf. 2014 Jul;13(4):377-399. doi: 10.1111/1541-4337.12065. PMID: 33412697
- Athane et al., The safety evaluation of phycocyaninenriched Galdieria sulphuraria extract using 90-day toxicity study in rats and in vitro genotoxicity studies. Toxicology Research and Application 2020.
- Minxi Wan et al., Comparison of C-phycocyanin from extremophilic Galdieria sulphuraria and Spirulina platensis on stability and antioxidant capacity, Algal Research 2021, Volume 58.
- Gross, W. & Schnarrenberger, C. (1995): Heterotrophic growth of two strains of the acido-thermophilic red alga Galdieria sulphuraria. Plant Cell Physiol. 36(4): 633-638.
- Niels T. Eriksen, Production of phycocyanin—a pigment with applications in biology, biotechnology, foods and medicine. Applied Microbiology and Biotechnology, 2008
Molecular Devices UK Ltd
Molecular Devices is one of the world’s leading providers of high-performance life science technology. We make advanced scientific discovery possible for academia, pharma, and biotech customers with platforms for high-throughput screening, genomic and cellular analysis, colony selection and microplate detection. From cancer to COVID-19, we've contributed to scientific breakthroughs described in over 230,000 peer-reviewed publications.
Over 160,000 of our innovative solutions are incorporated into laboratories worldwide, enabling scientists to improve productivity and effectiveness – ultimately accelerating research and the development of new therapeutics. Molecular Devices is headquartered in Silicon Valley, Calif., with best-in-class teams around the globe. Over 1,000 associates are guided by our diverse leadership team and female president that prioritize a culture of collaboration, engagement, diversity, and inclusion.
To learn more about how Molecular Devices helps fast-track scientific discovery, visit www.moleculardevices.com.
Sponsored Content Policy: AZO Life Sciences publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.