Three-dimensional (3D) organoids are gaining popularity throughout drug discovery and disease modeling as they can better represent biologically relevant microenvironments, functionality, and tissue architecture.
However, the complicated nature of 3D models continues to hinder their widespread adoption in research and drug screening.
Molecular Devices LLC has been investigating automation to help alleviate the hurdles of labor-intensive manual protocols. The company has developed a series of automation protocols for the 3D culture workflow, improving organoid assays’ throughput and accuracy.
These automated methods use an integrated work cell to offer automated cell plating, culture monitoring, media exchange, and high-content imaging. This novel method also enables the automation of compound addition, cell staining, and endpoint assays.
The company’s automated procedures can accommodate two of the most common organoid workflows:
- Organoids cultured in matrix (Matrigel); for example, primary tissue-derived, iPSC-derived intestinal or colorectal organoids
- Organoids cultured matrix-free using U shape ULA labware; for example, organoids used for spheroids and patient-derived breast cancer tumoroids
There are a range of benefits to this approach. In the examples presented in this article, processes for organoid assays were automated by integrating several instruments, enabling an automated culture of organoids, maintenance, compound treatment, and endpoint assays for different 3D cellular models.
This process facilitates the automation and scale-up of complex assays in 3D biology. These methods are also suitable for compound screening and disease modeling.
Materials and Methods
Cell Culture
Matossian et al. (2021) have previously outlined methods for generating tumoroids and PDX organoids (PDXO).
In this example, the primary tumor sample was implanted into SCID/Beige mice. This sample exhibited rapid tumor growth, taking 14 days to reach a maximal tumor volume greater than 1000 mm3.
A cell line was then generated from that sample, allowing it to be expanded in 2D culture. Tumoroids were created from 4IC cells expanded in 2D.
4IC cells were dispensed at a rate of approximately 2,000 cells per well in U-shape low-attachment 384 plates (Corning) before being incubated for 48 hours to form tight toroids.
These 4IC cells were cultured using Advanced DMEM supplemented with glucose, NEAA, 2 mM glutamine, and insulin 120 µg/L, 10 % FBS (Gibco 12491-015).
For metabolic assays, toroids were cultured with DMEM + 10 % dialyzed serum, 2 mM glutamine, 5 mM glucose, and no phenol red.
It was possible to derive 3D intestinal organoids from human iPSCs (STEMCELL Technologies) or primary mouse intestinal cells. The STEMCELL Technologies protocol informed the culturing and differentiation of cells, while IntestiCult™ Organoid Growth Medium (STEMCELL Technologies) was used for the cell culture.
Cells were seeded in 50 % growth-factor reduced Matrigel (Corning) domes in a 24-well plate format. These cells were fed with fresh media every second day for 7–10 days. Next, intestinal organoids were passaged-dissociated before being re-plated into fresh Matrigel domes.
It should be noted that this project was completed under the license from HuB Institute.
Cell Monitoring and Imaging
The ImageXpress Confocal HT.ai High-Content Imaging System (Molecular Devices) was used to acquire fluorescent images' transmitted light (TL). This was done using the MetaXpress High-Content Image Analysis Software.
Tumoroid images were acquired in TL using a roughly 60 µm offset, while Z-stack images were obtained using the 10x or 20x objectives in confocal mode. Analysis was conducted using the MetaXpress or IN Carta™ Image Analysis Software.
Automation of Cell Culture and Imaging Protocols
Automated imaging and analysis of organoids can greatly benefit the quantitative assessment of phenotypic changes in organoids and increase throughput in experiments and tests.
Molecular Devices LLC was able to construct an automated, integrated system to facilitate automated monitoring, maintenance, and characterization of the growth and differentiation of organoids and stem cells. This system was also able to test the impact of various compounds.
The automated system leveraged a robust combination of the ImageXpress Confocal HT.ai system and analysis software, a Biomek i7 or Hamilton liquid handler, an automated CO2 incubator, and a collaborative robot and rail. The Green Button Go solution enabled robotic automation.

Figure 1. The integrated system includes an automated CO2 incubator, liquid handler, IXM-C HT.ai confocal imaging system, automation compatible plate reader and centrifuge A Collaborative robot and scheduling software allows automated plate transfer between instruments, and scheduling of different steps. Image Credit: Molecular Devices UK Ltd
Results
Automated Culturing 3D Organoids in Matrigel
Culturing 3D organoids in Matrigel or other matrices includes several steps, including seeding primary or iPSC-derived cells mixed with Martigel into the “domes” and a further 8-10 days of culture with media exchange every other day.
Automated imaging in transmitted light was used to monitor the growth and maturation of organoids. Organoids were harvested following the re-suspension of domes, and these were disintegrated using either repeated mechanical pipetting or enzymatic treatment.
Cell pellets were then centrifuged before being mixed with Matrigel and re-plated onto new plates.
An efficient automated liquid handling system enabled the automated seeding of colorectal, intestinal, or other cells in Matrigel droplets into several different plate formats, as well as automated media exchange, passaging, and compound addition.
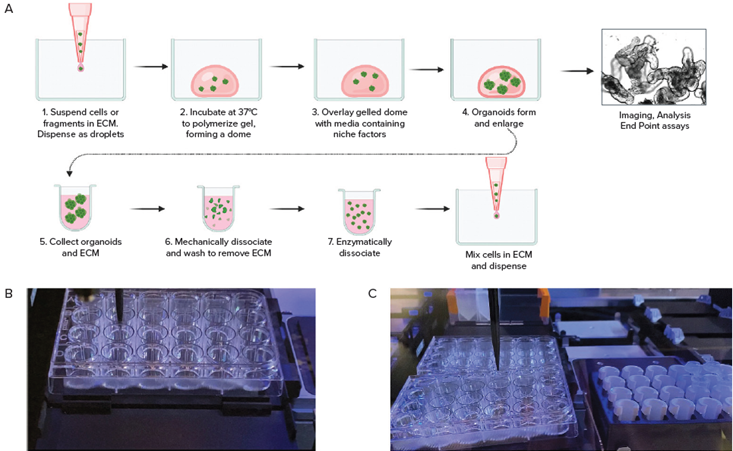
Figure 2. A. Schematic diagram of organoid culture workflow. B. Automated seeding organoids in domes; C. Cell collection by Hamilton liquid handling system. Image Credit: Molecular Devices UK Ltd
Monitoring Development of 3D Organoids and High Content Imaging Assays
Automated imaging in transmitted light was employed to monitor the development of organoids while organoids were located, and their size and complexity were defined using InCarta SW.
Notably, organoids grown in Matrigel will self-organize and develop complex structures, lumens, and crypts. Machine learning-based image analysis can accurately measure organoid diameter, size, and density and characterize their complexity by counting crypts or lumens.
Organoids were stained for differentiation or viability markers to enable endpoint measurements. Automated confocal imaging and image analysis enabled the complex phenotypic evaluation of organoid structures in three dimensions.
Organoid phenotypes were characterized according to the number of organoids, their size distribution, and complexity (number of crypts or cavities). The cell content per organoid was also investigated alongside their cell viability, nuclei, mitochondria, and cytoskeleton.
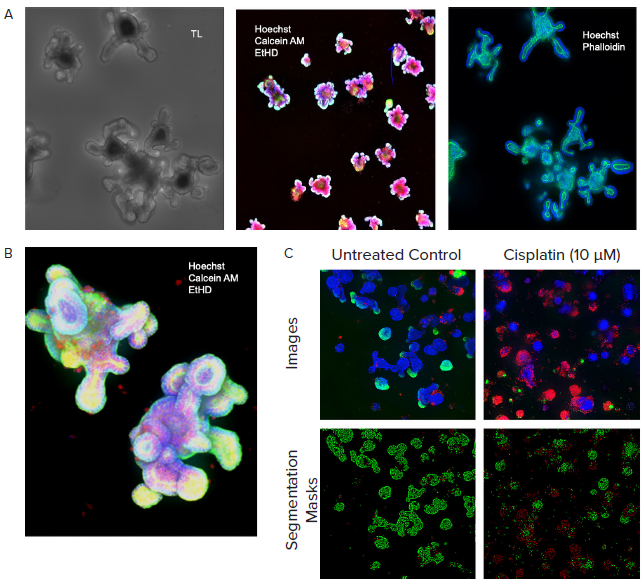
Figure 3. A. Monitored organoid development in transmitted light; live staining of organoids with viability dyes; fixed organoids stained with phalloidin. B. Organoids cultured for 10 days, stained with viability dyes. C. Control and treated organoids stained with viability dyes, live-dead analysis provided by MetaXpress analysis software. Image Credit: Molecular Devices UK Ltd
Culturing and Imaging 3D Tumoroids
A number of steps are generally required when culturing 3D spheroids or tumoroids matrix-free to enable the formation of spheroids in U-shape low-attachment plates.
By automating liquid exchange and compound addition, it is possible to work with non-attached microtissue cultures without risking breaking or losing them during processing.
In the example presented here, 3D tumoroid culture was created from a primary triple-negative tumor; while the cell line was developed by passaging primary tissues in SCID mice prior to adapting these for 2D cell culture.
Tumoroids were formed by culturing 2,000 cells in 384 well-low attachment plates for 48 hours. These tumoroids were then treated with compounds from the library of approved anti-cancer drugs provided by the National Cancer Institute (NCI).
Five concentrations were used for testing, while Biomek automation was utilized in compound dilutions, cell treatments, and staining. Tumoroids were then cultured and subjected to daily monitoring via automated imaging.
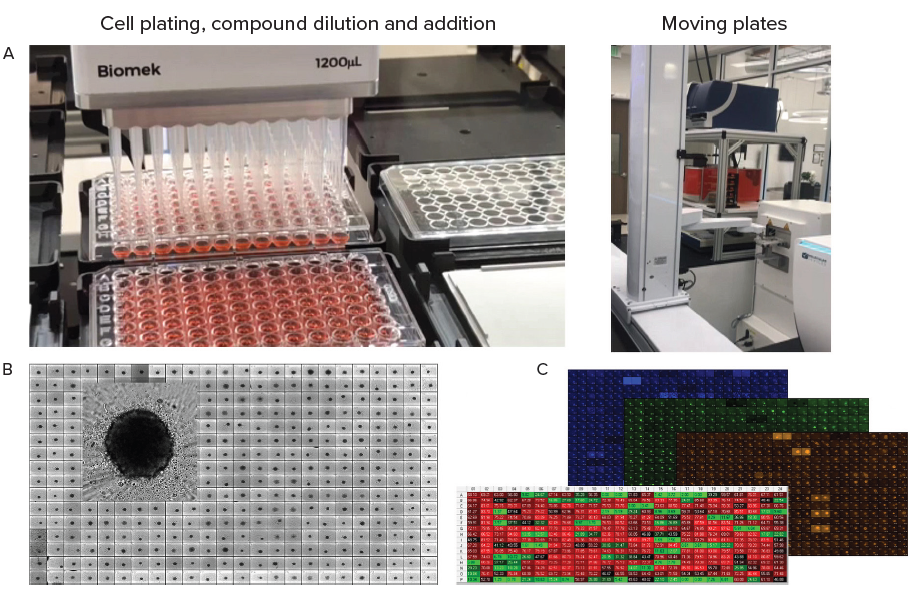
Figure 4. A. Automation of organoid culture, compound addition and staining. B. Tumoroids formed 48h after plating, TL images (10X). C. Tumoroids were treated with compounds for 5 days then stained with Hoechst dye (blue), Calcein AM (green) and EtHD (red), 10X. Organoids were imaged using confocal option, Z-stack of 15 images 10 μm apart,. Image analysis was done using Custom Module Editor (MetaXpress software) for finding organoids, nuclei, live and dead cells. Image Credit: Molecular Devices UK Ltd
Automation of Cell Culture, Compound Treatment, and Imaging
During the endpoint assay, the cells under investigation were stained with a combination of Calcein AM, Hoechst, and EtHD before analysis via the Custom Module Editor for complex phenotypic analysis.
AI-based image analysis was used to automatically detect and characterize tumoroid phenotypes, density, and sizes.
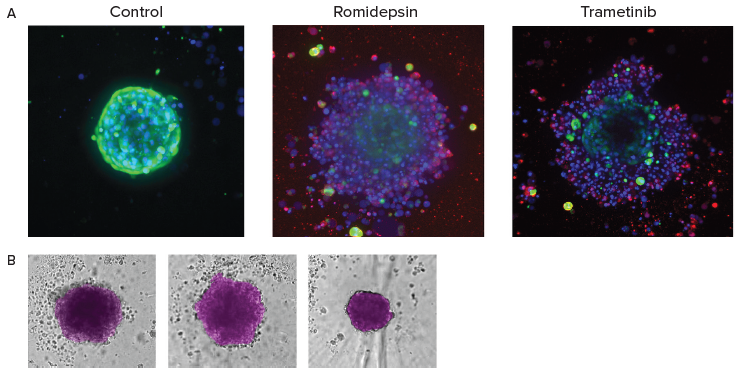
Figure 5. A. End-point analysis of fluorescent images was done using Custom Module Editor ImageXpress software. Images (maximum projection), and analysis masks shown. Multiple measurements were derived for cell scoring and organoid characterization. B. Automated monitoring and image analysis of 3D cancer microtissues was done using transmitted light images (10X) with AI-based image analysis In Carta software (analysis mask shown in purple). Image Credit: Molecular Devices UK Ltd
Conclusions
In the example presented here, organoid assays and compound treatment processes were automated by integrating several instruments. This combination of instruments enabled automated cell culture, maintenance, and compound treatment of 3D cellular models.
It is possible to employ this process in automation and compound screening in 3D biology.
It has been shown that a combination of confocal imaging and 3D analysis enables the complex, quantitative analysis of cellular content of organoids and the count and measurements of cells with various phenotypes.
The methods outlined in this article can be employed to evaluate phenotypic changes triggered by test compounds and disease modeling.
Acknowledgments
Produced from materials originally authored by Oksana Sirenko, Zhisong Tong, Krishna Macha, and Angeline Lim from Molecular Devices LLC.
About Molecular Devices UK Ltd
Molecular Devices is one of the world’s leading providers of high-performance life science technology. We make advanced scientific discovery possible for academia, pharma, and biotech customers with platforms for high-throughput screening, genomic and cellular analysis, colony selection and microplate detection. From cancer to COVID-19, we've contributed to scientific breakthroughs described in over 230,000 peer-reviewed publications.
Over 160,000 of our innovative solutions are incorporated into laboratories worldwide, enabling scientists to improve productivity and effectiveness – ultimately accelerating research and the development of new therapeutics. Molecular Devices is headquartered in Silicon Valley, Calif., with best-in-class teams around the globe. Over 1,000 associates are guided by our diverse leadership team and female president that prioritize a culture of collaboration, engagement, diversity, and inclusion.
To learn more about how Molecular Devices helps fast-track scientific discovery, visit www.moleculardevices.com.
Sponsored Content Policy: AZO Life Sciences publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.