Organoids derived from patients (PDOs), including those sourced from tumors, are utilized as predictive models to gauge patient responses to drugs and treatments. The integration of the BioAssembly™ platform has proven effective in enhancing the intricate and essential three-dimensional (3D) tissue models for drug screening purposes.
This cutting-edge automation solution seamlessly incorporates the BioAssemblyBot® (BAB), BioStorageBot™ modular incubator, and BioApps™ intuitive workflow control software with confocal imaging into the workflow, enhancing agility and efficiency.
After automated preparation, feeding, drug treatment, and image acquisitions, image sets were analyzed to produce comprehensive assessments of PDO responses to the drugs, yielding high-content insights.
The BAB platform enhances the precision, efficiency, and reproducibility of dispensing PDOs into multiwell plates through the utilization of finely adjustable operational parameters, such as pipetting speed, thereby reducing organoid fragmentation and ensuring uniform plating.
In the study discussed in this article, an automated PDO drug screen involving automated imaging and High Content Analysis (HCA) with the BAB platform was performed.
Figure 1 illustrates the workflow, which entailed evaluating the response of colorectal cancer PDOs sourced from two distinct patients to the drugs trametinib and adavosertib across four varying doses.
A total of 76 image features were extracted from the image sets generated during the automated workflows, which were subsequently assessed for drug- and dose-specific responses. The automated PDO assay with HCA successfully demonstrated its ability to distinguish between PDOs treated with different drugs.
Benefits of PDO and Workflow Automation
- Applicable to a variety of PDO types and systems.
- Automation enhances the quality, throughput, and consistency of assays.
- BioAssembly’s flexible and easy-to-use platform is conducive to automated imaging and high-content analysis.
- Easy-to-use BioApps software integrates and controls all components.
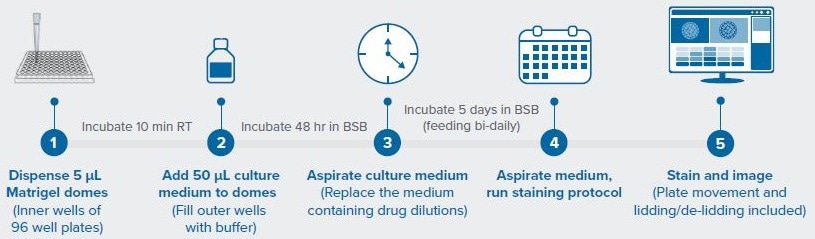
Figure 1. Workflow for automated PDO drug screen. Image Credit: Molecular Devices UK Ltd
Materials and Methods
Colorectal cancer (CRC) PDOs from two different donors were supplied by Molecular Devices. These PDOs were suspended in 80% Matrigel® and then dispensed into a 96-well plate using the BAB and pipette hand. A dome was created by dispensing 5 µl into the center of each well. After gelling, the BAB introduced culture medium into each well and subsequently transferred the plate to the modular BioStorageBot incubator.
On the second day of culture, the BAB changed culture media and introduced a serial dilution series of trametinib (MEK1/2 inhibitor) or adavosertib (tyrosine kinase WEE1 inhibitor) to the wells.
After seven days, cultures were imaged using an integrated confocal imager, following a Hoechst staining protocol. Offline, image analysis using IN Carta software was performed on the maximum projection images. Multiparametric analyses were also carried out using StratoMineR from Core Life Analytics.
Following imaging, a CellTiter-Glo® 3D assay was performed to measure Adenosine triphosphate (ATP) production. All steps of the PDO assay, including the automated imaging protocol, were controlled by a user-defined PDO BioApp software module.
Results and Discussion
Properly dispensing PDOs in Matrigel can be challenging as to form a dome, it must be deposited exactly in the center of the well. The dome is necessary to prevent Matrigel from forming a meniscus at the well edge, which can complicate imaging.
If carried out manually, quite a bit of practice would required if the hope is to achieve a high success rate of dome formation. However, the incorporation of the BAB has proven to improve the dome formation rate by 10 % and reduce dome dispensing time by 50 %, compared with no automation, as shown in Figure 2.
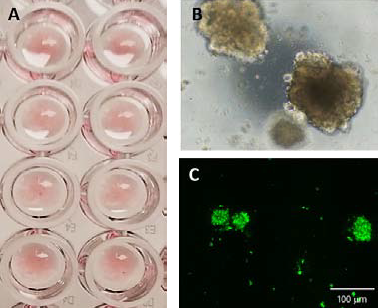
Figure 2. CRC PDOs seeded in Matrigel domes in a 96 well plate (A). PDOs after 7 days of culture (B) phase contrast image, and (C) calcein AM. Image Credit: Molecular Devices UK Ltd
After two days, serial dilutions of trametinib and adavosertib were introduced to wells. The PDOs were incubated for an additional five days, after which an end-point viability assay was performed.
CellTiter-Glo 3D, a tool for measuring ATP production as an indirect measure of cell viability, demonstrated apparent differences between the highest and lowest drug concentrations and the untreated controls, as illustrated in Figure 3. Despite this, no discernable change in organoid diameter in response to the inhibitors was observed (Figure 4).
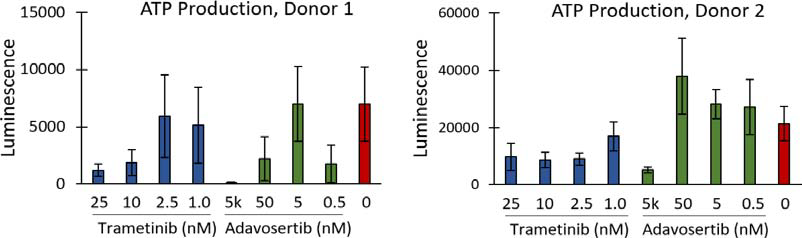
Figure 3. ATP production of PDOs, as measured by CellTiter-Glo 3D, performed at the end of the assay. Image Credit: Molecular Devices UK Ltd
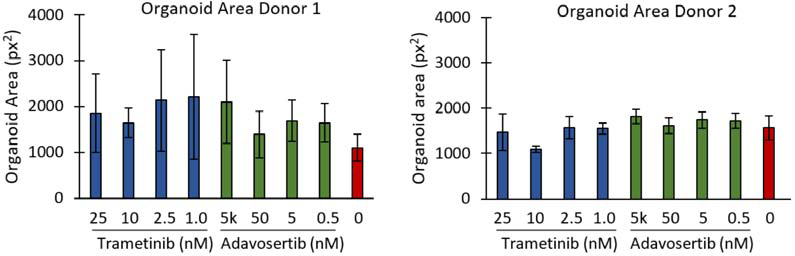
Figure 4. Area of PDOs treated with different concentrations of trametinib or adavosertib, compared to untreated controls. Plates were seeded with PDOs from one of two different donors. Bars are mean ± SD. Image Credit: Molecular Devices UK Ltd
Deep-learning-based segmentation in IN Carta was utilized to extract features such as area, intensity, form factor, and texture from the data set. These features were then examined using StratoMineR’s cloud-based advanced analytics.
This study focused on principal component analysis (PCA) and Euclidian distance analyses as baseline readouts for the automated workflow. PCA of up to 76 features identified differential responses by the PDOs to the inhibitors, as shown in Figure 5.
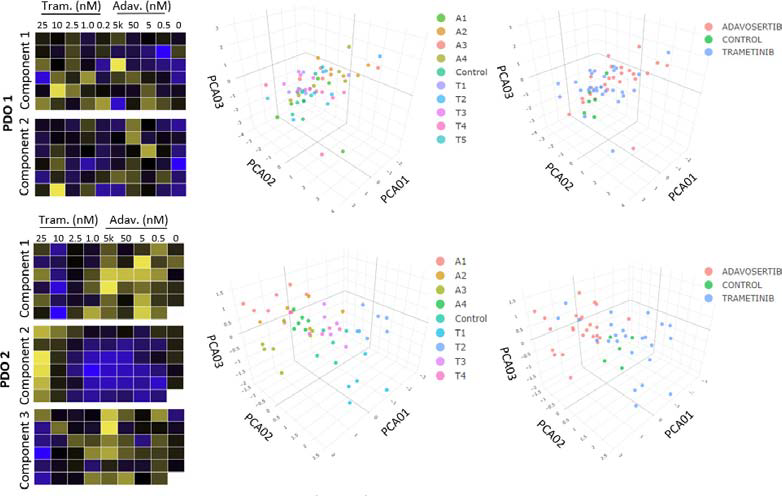
Figure 5. Heat maps and principal component analysis (PCA). For Donor 1 (PDO 1), 2 components are shown, which show limited clustering within or between drug concentrations. Donor 2, however, had clear clustering in components 1, 2, and 3, within each drug compared to controls. Image Credit: Molecular Devices UK Ltd
PDOs from Donor 1 were shown to have limited response to the two inhibitors, while PDOs from Donor 2 displayed clear differences in inhibitor responses in a dose-dependent manner.
Assessment of the hit rate through phenotypic distance mapping confirmed the relatively low responsiveness of Donor 1 PDOs to the inhibitors, with a hit rate of 31.5 %, as illustrated in Figure 6. Conversely, both inhibitors substantially impacted Donor 2 PDOs, with a hit rate of 100 %.
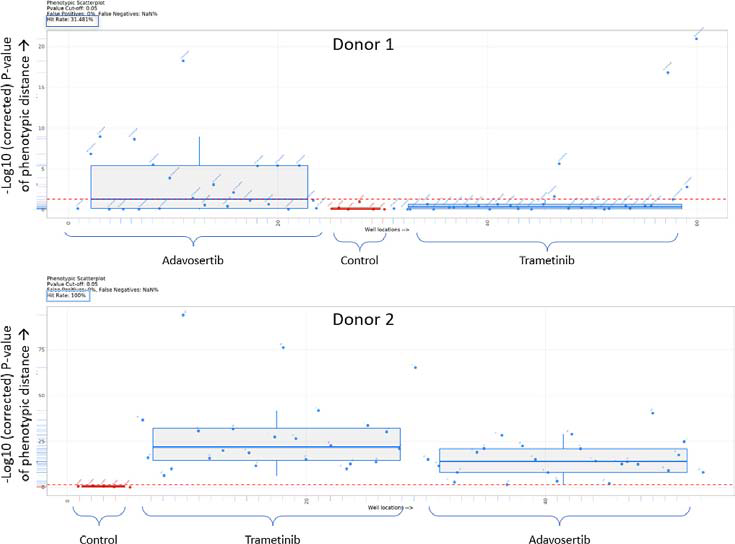
Figure 6. Box plots showing the Euclidian distance from the median of the control (untreated) for PDO 1 and PDO 2. Everything above the red dotted line has a p value <0.05. Image Credit: Molecular Devices UK Ltd
Conclusions
This study showcases the efficacy of a fully integrated automation platform for screening the impacts of therapeutic compounds on patient-derived tumor organoids. The automated PDO assay successfully detected patient-specific variations in PDO responses to selected signaling inhibitors through high-content image analyses.
Although further research is still required to establish measurements of consistency, accuracy and precision, the automated workflow outlined in this article was capable of producing results comparable to those obtained by a skilled scientist in approximately half the time and with improved efficiencies.
Combining automated PDO formatting, culturing, treating and imaging with high-content analysis represents a powerful, simple and highly adaptable method for high-throughput drug screening.
Acknowledgments
Produced from materials originally authored by Hannah A. Strobel from Advanced Solutions Life Sciences, Victoria Marsh and Angeline Lim from Molecular Devices, and Maria R. Oyaga from Core Life Analytics.
About Molecular Devices UK Ltd 
Molecular Devices is one of the world’s leading providers of high-performance life science technology. We make advanced scientific discovery possible for academia, pharma, and biotech customers with platforms for high-throughput screening, genomic and cellular analysis, colony selection and microplate detection. From cancer to COVID-19, we've contributed to scientific breakthroughs described in over 230,000 peer-reviewed publications.
Over 160,000 of our innovative solutions are incorporated into laboratories worldwide, enabling scientists to improve productivity and effectiveness – ultimately accelerating research and the development of new therapeutics. Molecular Devices is headquartered in Silicon Valley, Calif., with best-in-class teams around the globe. Over 1,000 associates are guided by our diverse leadership team and female president that prioritize a culture of collaboration, engagement, diversity, and inclusion.
To learn more about how Molecular Devices helps fast-track scientific discovery, visit www.moleculardevices.com.
Sponsored Content Policy: AZO Life Sciences publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.