Reviewed by Lauren HardakerJan 19 2026
Humanized mouse models are essential for investigating human gene function; however, the complete replacement of mouse genes with entire human sequences has posed significant technical challenges.
 Image credit: ImageFlow/Shutterstock.com
Image credit: ImageFlow/Shutterstock.com
The researchers have created a streamlined two-step CRISPR technique that successfully inserts substantial human genomic regions into mouse embryonic stem cells. The resulting mice exhibited human-like splicing, tissue-specific expression, and normal biological functions. This method also facilitates the incorporation of disease-related mutations, providing a flexible platform for generating precise, physiologically relevant humanized models.
Understanding human gene function in living organisms has historically been hindered by fundamental interspecies differences. While mice possess the majority of protein-coding genes found in humans, their regulatory landscapes frequently differ, which restricts the accuracy with which mouse models can replicate human biology.
A promising approach to address this issue is full-length gene humanization (FL-GH), where entire mouse loci, including coding sequences, introns, untranslated regions, and regulatory elements, are substituted with their human equivalents. The current technologies have faced challenges in efficiently and reliably inserting very large genomic fragments, hindering the progress towards developing physiologically relevant humanized models.
The research team, from The Institute of Medical Science at The University of Tokyo, Japan, has created an efficient two-step approach for FL-GH. The study was published in Nature Communications. The study presents TECHNO (Two-step ES Cell-based HumaNizatiOn), a technique that combines CRISPR/Cas9-assisted genome editing with the delivery of large human genomic regions via bacterial artificial chromosomes (BAC). This framework provides a viable, scalable method for substituting entire mouse loci with their human counterparts.
Our results demonstrate a robust and broadly applicable platform for generating FL-GH mouse models.
Dr. Manabu Ozawa, Study Head and Associate Professor, The Institute of Medical Science, The University of Tokyo, Japan
The TECHNO workflow is executed in two synchronized phases. Initially, the target mouse locus is removed utilizing Cas9 ribonucleoproteins and substituted with short human homology arms that encase a selection cassette, thereby establishing a precise genomic landing site. In the subsequent phase, a BAC containing the complete human gene along with its regulatory elements is introduced into embryonic stem cells, accompanied by a universal gRNA that targets the selection cassette, facilitating homology-directed integration of genomic fragments that exceed 200 kbp. Given that this method depends on standard molecular reagents and readily accessible BAC libraries, it is theoretically applicable to over 90 % of human genes.
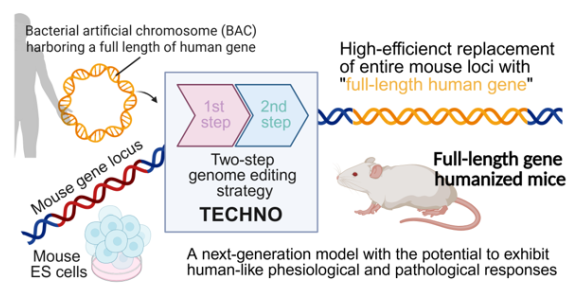 TECHNO: A two-step genome editing strategy that enables efficient replacement of entire mouse loci with full-length human genes to generate gene-humanized mice. Image Credit: University of Tokyo
TECHNO: A two-step genome editing strategy that enables efficient replacement of entire mouse loci with full-length human genes to generate gene-humanized mice. Image Credit: University of Tokyo
Through this platform, the team successfully humanized multiple loci, including c-Kit, APOBEC3, and CYBB. The humanization of c-Kit replicated human-like alternative splicing and organ-specific expression while supporting critical biological functions such as hematopoiesis and spermatogenesis.
The replacement of the APOBEC3 locus illustrated the scalability of the method, integrating more than 200 kbp of human DNA that encompasses seven genes and producing expression patterns that reflected those seen in humans.
The researchers developed a humanized CYBB allele and incorporated disease-associated mutations to model chronic granulomatous disease. The resulting mice exhibited reduced reactive oxygen species production, accurately mirroring the molecular phenotype observed in patients.
In the short term, TECHNO is anticipated to expedite the creation of accurate, human-relevant animal models for assessing therapeutic targets, confirming disease-related variants, and recognizing ineffective drug candidates sooner in research pipelines.
In the long run, scalable FL-GH has the potential to transform biomedical research by facilitating models that more accurately replicate human gene regulation and disease processes.
These developments also pave the way for the incorporation of humanized models into AI-enhanced comparative genomics, extensive humanized allele panels, and systems biology frameworks.
Overall, these results demonstrate that our method enables not only FL-GH of individual loci but also precise modeling of human genetic diseases in vivo by introducing disease-associated mutations into humanized alleles.
Dr. Manabu Ozawa, Study Head and Associate Professor, The Institute of Medical Science, The University of Tokyo, Japan
The TECHNO platform signifies a significant progression towards next-generation humanized mouse models by facilitating the stable and high-efficiency integration of genomic fragments larger than 200 kbp, all while maintaining intricate regulatory behaviors in vivo.
Its adaptability, durability, and dependence on conventional laboratory instruments establish it as a cornerstone technology for the advancement of functional genomics, disease modeling, and translational medicine.
Source:
Journal reference:
Taguchi, J., et al. (2026) A scalable two-step genome editing strategy for generating full-length gene-humanized mice at diverse genomic loci. Nature Communications. DOI: 10.1038/s41467-025-67900-4. https://www.nature.com/articles/s41467-025-67900-4