Researchers from the University of Illinois Chicago have created software that can aid researchers in identifying gene regulators faster. To anticipate which transcription factors are most likely to be active in individual cells, the system uses a machine-learning algorithm.
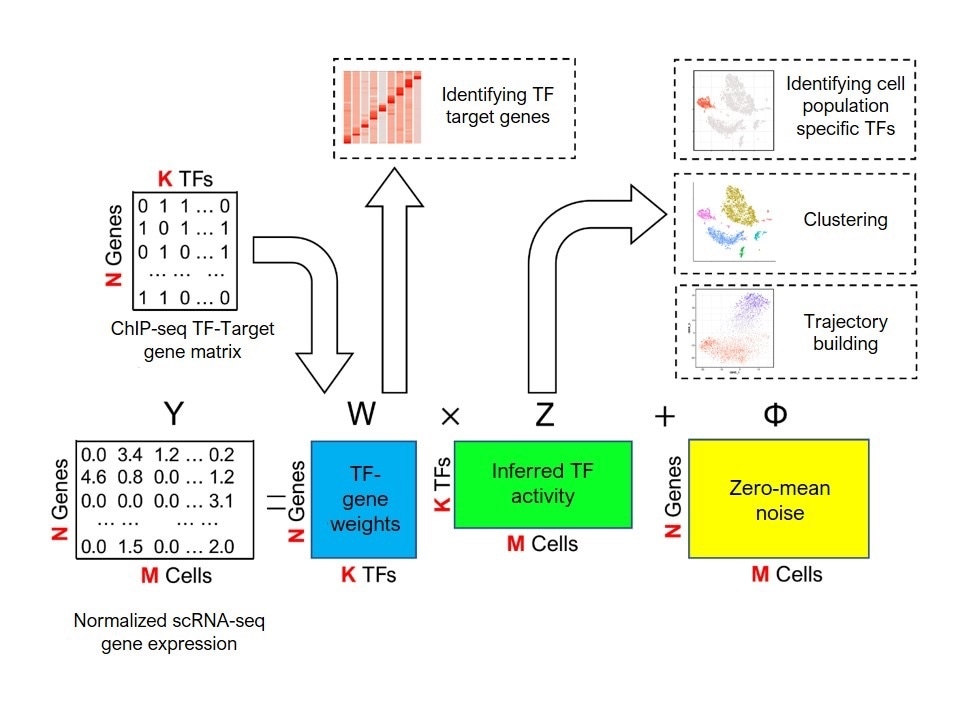
A schematic overview of the BITFAM machine learning system developed by researchers at UIC. User-provided sequencing data (“Normalized scRNA-Seq gene expression”) and existing data on transcription factor binding sites (“ChIP-seq TF-Target gene matrix”) are analyzed to predict transcription factor activity (“Inferred TF activity”) that can be leveraged for a broad range of analyses. Image Credit: Genome Research, Attribution 4.0 International CC BY 4.0 license.
Transcription factors are proteins that bind to DNA and regulate which genes in a cell are turned on or off. Understanding and controlling these signals in the cell can be an effective method to identify new treatments for some ailments; therefore, these proteins are important to biomedical researchers.
But hundreds of transcription factors exist inside human cells and determining which are the most active—those that are expressed, or “on”—in different types of cells and could be used as medication targets can take years of research, often through trial and error.
One of the challenges in the field is that the same genes may be turned “on” in one group of cells but turned “off” in a different group of cells within the same organ. Being able to understand the activity of transcription factors in individual cells would allow researchers to study activity profiles in all the major cell types of major organs such as the heart, brain or lungs.”
Jalees Rehman, Professor, Department of Medicine and Department of Pharmacology and Regenerative Medicine, College of Medicine, University of Illinois Chicago
The UIC-developed method, termed BITFAM (Bayesian Inference Transcription Factor Activity Model), works by merging fresh gene expression profile data from single-cell RNA sequencing with existing biology data on transcription factor target genes.
The system uses this data to run a series of computer-based simulations to discover the best fit and predict the activity of each transcription factor in the cell.
The method was tested in cells from lung, heart, and brain tissue by UIC researchers led by Rehman and Yang Dai, a UIC associate professor in the department of bioengineering in the Colleges of Medicine and Engineering. The model’s details, as well as the results of their experiments, were published in the journal Genome Research.
Our approach not only identifies meaningful transcription factor activities but also provides valuable insights into underlying transcription factor regulatory mechanisms. For example, if 80% of a specific transcription factor’s targets are turned on inside the cell, that tells us that its activity is high.”
Shang Gao, Study First Author and Doctoral Student, Department of Bioengineering, University of Illinois Chicago
“By providing data like this for every transcription factor in the cell, the model can give researchers a good idea of which ones to look at first when exploring new drug targets to work on that type of cell,” added Gao.
The new approach, according to the researchers, is open source and might be used widely since users can combine it with other analysis methods that are better suited for their investigations, such as finding new drug targets.
This new approach could be used to develop key biological hypotheses regarding the regulatory transcription factors in cells related to a broad range of scientific hypotheses and topics. It will allow us to derive insights into the biological functions of cells from many tissues.”
Yang Dai, Associate Professor, Department of Bioengineering, College of Medicine and College of Engineering, University of Illinois Chicago
According to Rehman, whose research focuses on the processes of inflammation in vascular systems, the new technique might be used to focus on the transcription factors that drive diseases in specific cell types, which is something his team is interested in.
“For example, we would like to understand if there is transcription factor activity that distinguished a healthy immune cell response from an unhealthy one, as in the case of conditions such as COVID-19, heart disease or Alzheimer’s disease where there is often an imbalance between healthy and unhealthy immune responses,” concluded Dai.
Source:
Journal reference:
Gao, S., et al. (2021) A Bayesian inference transcription factor activity model for the analysis of single-cell transcriptomes. Genome Research. doi.org/10.1101/gr.265595.120.