Reviewed by Lauren HardakerFeb 6 2026
Proteins are the molecular machines of cells. They are manufactured in protein factories known as ribosomes using their blueprint, or genetic information. Here, amino acids, the fundamental building blocks of proteins, are organized into lengthy protein chains. Individual proteins, like the building pieces of a machine, require a precise three-dimensional shape to operate properly.
 Image credit: Christoph Burgstedt/Shutterstock.com
Image credit: Christoph Burgstedt/Shutterstock.com
To do this, freshly formed protein chains in human cells are folded into stable, functional forms with the assistance of several protein-folding chaperones, such as TRiC/PFD or HSP70/40. Protein folding chaperones help separate amino acid chains from the cellular environment, which have varied chemical characteristics depending on the amino acid. This prevents freshly formed protein chains from aggregating and causing disease.
F.-Ulrich Hartl, a director at the Max Planck Institute of Biochemistry, has spent decades researching the mechanisms behind protein folding.
Much of what we know about protein folding has been learned from studies conducted in test tubes. However, it is virtually impossible to faithfully replicate the cellular environment in vitro. Unlike a test tube, a cell is a highly complex environment filled with many different macromolecules, like proteins, nucleic acids and lipids.
Niko Dalheimer, Study Co-Lead Author and Scientist, Max Planck Institute
Dalheimer added, “To fully understand how chaperones work, we examined the protein folding dynamics of TRiC and PFD in its natural environment – intact human cells – using single-particle tracking on a fluorescence microscope, an approach that has only recently become feasible, thanks to advances in live-cell fluorescent labeling.”
TRiC and Prefoldin
Through a channel, the ribosomes progressively release a freshly made protein as an amino acid chain. Prefoldin, or PFD for short, is a co-chaperone of TRiC that captures and protects the free amino acid residues to keep the freshly generated protein from clumping together. The protein is subsequently transferred to the chaperonin TRiC for folding by the co-chaperone.
TRiC, a barrel-shaped protein-folding aid, is associated with the bacterial GroEL/ES.
Although TRiC only helps 10 % of the proteins in a cell to fold, many of them are particularly important for the cell, including actin and tubulin, which are building blocks of the cytoskeleton. That's why we looked at this part of protein folding. We used actin as a test protein to understand the folding dynamics in cells.
Rongqin Li, Study Co-First Author and Scientist, Max Planck Institute
Single Particle Tracking Sheds Light on the Unknown
To track the real-time interaction of all components involved in protein folding, the researchers labeled TRiC and prefoldin, as well as the actin nascent chain, as direct chaperone substrates and ribosomes and mRNAs as chaperone substrate proxies in various settings with green and magenta colors.
If the two components were close together, fewer than 500 nanometers apart, the colors merged and appeared as white dots under the microscope.
There are approximately 10 million ribosomes in a single cell. To enable us to track individual ribosomes and other components under the microscope, we stained only a small proportion of the ribosomes, rather than all of them and used the TIRF method to track the individual molecules and their interactions with chaperones. It is like a diver exploring a pitch-dark deep sea: by shining a light on just a few spots at a time, the diver can get a glimpse of the hidden dynamic life and activity around them.
Niko Dalheimer, Study Co-Lead Author and Scientist, Max Planck Institute
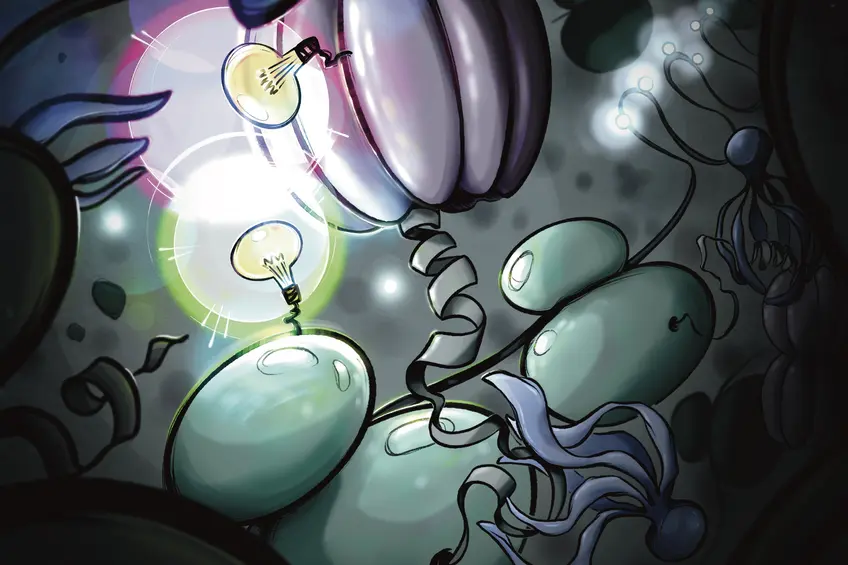
Artistic view of a living human cell, where sparse fluorescent labels, depicted as light bulbs, illuminate the TRiC chaperonin and ribosome amid molecular complexity. A newly synthesized protein emerging from the ribosome engages with the squid-like prefoldin and is delivered to the barrel-shaped TRiC chaperonin. This method enables real-time single-molecule insight into how proteins are guided along their folding pathway inside the cell. Image Credit: MPI of Biochemistry/ Marzia Munafo
Repeated Contact
TRiC and PFD probed the freshly generated actin protein chain coming from the ribosome for around one second each, according to the researchers. PFD holds the nacent chain immediately before actin is released from the ribosome and transfers it to the TRiC champer for folding completion.
“Interestingly, the contact between TRiC and actin mutants, i.e., protein chains into which we introduce errors to disrupt its proper folding, was significantly longer. In contrast to the normal condition, the folding-defective actin undergoes multiple-rounds of attempted folding by the chaperonin system and is ultimately targeted for degradation,” Rongqin Li added.
F.-Ulrich Hartl summarized, “For decades, we and others have studied chaperone-mediated protein folding primarily through biochemical experiments, which have been essential for defining how this process is controlled. With live-cell single-particle tracking, we can now examine these concepts directly in living cells.”
Hartl concluded, “In doing so, we have confirmed key findings from classical biochemical experiments, while at the same time uncovering features – such as the protected folding zone – that could not have been detected with ensemble-based assays. This is the first time these processes have been visualized at the single-molecule level in living cells. As I often tell my colleagues, ‘seeing is believing.”
Source:
Journal reference:
Li, R. et.al. (2026) Single-molecule dynamics of the TRiC chaperonin system in vivo. Nature. DOI: 10.1038/s41586-025-10073-3. https://www.nature.com/articles/s41586-025-10073-3.