Unexpected heterogeneity of ribosomal RNA genes in human populations revealed by genome studies suggests potential variation in protein translation by the ribosomes. Image Credit: Wenjun Fan, PhD
Human ribosomal RNA (rRNA) genes are required for the formation of ribosomes, or protein-translating machines. The findings of the study showed that these genes, which were previously assumed to be comparable among people, changed dramatically depending on a person’s geographic heritage. The results were published on February 2nd, 2022, in the journal RNA.
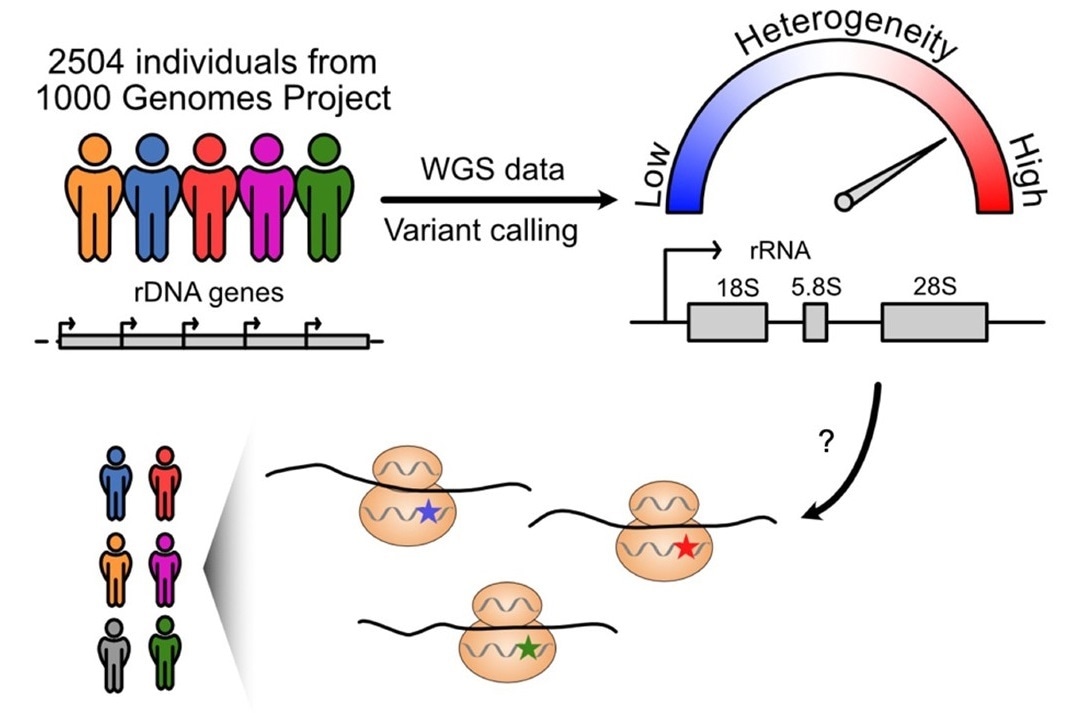
A section termed 28S rRNA, a key component of the protein-translating ribosome, has particularly significant variability.
The team, headed by Marikki Laiho MD, PhD, director of molecular radiation sciences in the Department of Radiation Oncology and Molecular Radiation Sciences, differs from their usual research focus of developing new molecules that could be useful in cancer treatment to examine a basic biology concept for better understanding.
Researchers had created cancer medications that target ribosomal rRNA synthesis, a unique mechanism that causes cancer growth. Cancer cells cannot proliferate without them. The researchers questioned if malignancies changed the rRNA gene, and if so, how that may influence their targeting technique. Despite the gene’s relevance, there has yet to be a reliable reference sequence released.
Team members used high-performance computers at the Maryland Advanced Research Computing Center, a joint venture administered by The Johns Hopkins University and the University of Maryland, to adopt a bioinformatics approach to rRNA gene sequences.
Researchers needed to know whether there were any variances in the human population before they could start documenting cancer changes. The rRNA gene sequence was thought to be “untouchable,” or so basic that having any alteration is unlikely.
However, when we started that analysis, we very quickly realized that the cancer genomes were highly aberrant. In order for us to understand whether that aberration is real—meaning that it changes in a particular cancer—we needed to better understand what a normal human gene looks like.”
Marikki Laiho, Director, Molecular Radiation Sciences, Department of Radiation Oncology and Molecular Radiation Sciences, Johns Hopkins University
Researchers next analyzed variations in 2,504 people from 26 populations using whole-genome sequencing data from the 1000 Genomes Project (an international human genetics database). On the rRNA gene, they discovered 3,791 variation locations.
On 28S rRNA, there were 470 variant locations. The majority of these variations were found on large projecting folds of rRNA that differed between species. These are diverse positions that are expected to continuously evolve over time.
The analysis results were beyond our imagination. We saw perfect conservation of sequences across vast swaths of the gene, and then highly variable sites in the exact locations that we expected to be unaltered. This suggests that the way variant rRNAs are built into the ribosomes could lead to potential alterations in how the ribosomes function.”
Marikki Laiho, Director, Molecular Radiation Sciences, Department of Radiation Oncology and Molecular Radiation Sciences, Johns Hopkins University
Many of the detected variations were found to be separated by population. For instance, some variations were significantly more common in African or Asian groups than in American or European ones, and vice versa. According to Laiho, this raises the likelihood that some of the mutations are old and ancestry-dependent, yet have survived in present populations.
It’s too early to speculate what these variants mean, but what is fascinating is that they are conserved by population, and this means their retention is somehow important.”
Marikki Laiho, Director, Molecular Radiation Sciences, Department of Radiation Oncology and Molecular Radiation Sciences, Johns Hopkins University
The findings imply that a functional assessment of how the 28S rRNA variations alter ribosome activities is needed, which might lead to more specific cancer or other disease therapies, Laiho concluded.